 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aradu.F8RAG | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 295aa MW: 33654.7 Da PI: 5.4773 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 57.9 | 2.3e-18 | 23 | 70 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd++l+ +++++G g+W++ ++ g+ R++k+c++rw +yl
Aradu.F8RAG 23 KGPWTAEEDQILISYIQKHGHGNWRALPKQAGLLRCGKSCRLRWINYL 70
79******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 52.1 | 1.5e-16 | 76 | 121 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T eE+e +++++++lG++ W++Ia++++ gRt++++k+ w+++l
Aradu.F8RAG 76 RGNFTIEEEETIIKLHEMLGNR-WSAIAAKLP-GRTDNEIKNVWHTHL 121
89********************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 8.5E-27 | 14 | 73 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 18.143 | 18 | 70 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 5.08E-32 | 19 | 117 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.2E-15 | 22 | 72 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.3E-17 | 23 | 70 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.21E-11 | 25 | 70 | No hit | No description |
| PROSITE profile | PS51294 | 25.739 | 71 | 125 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.6E-26 | 74 | 126 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.5E-13 | 75 | 123 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.2E-15 | 76 | 121 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.16E-9 | 78 | 121 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 295 aa Download sequence Send to blast |
QHKKLQEAKM VRAPCCEKMG LKKGPWTAEE DQILISYIQK HGHGNWRALP KQAGLLRCGK 60 SCRLRWINYL RPDIKRGNFT IEEEETIIKL HEMLGNRWSA IAAKLPGRTD NEIKNVWHTH 120 LKKRLIKPNQ MNSSSSDTKR VSNKSKIKRS DSNSSTITVQ SSEPASLNSN FPEVDTTSAS 180 CTTTTTTTSS EFSSATNTGE RKIMNMMNIK NEELEDSLEN IIDESLWTEA GIDDETTNMT 240 ISNNELPSLQ YYDPVFNNSE ENLCFQPSAN FEDDGMDFWY DIFIRTGESI ELPEF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 5e-27 | 21 | 125 | 5 | 108 | B-MYB |
| 1h8a_C | 5e-27 | 21 | 125 | 25 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in cold-regulation of CBF genes and in the development of freezing tolerance. May be part of a complex network of transcription factors controlling the expression of CBF genes and other genes in response to cold stress. Binds to the MYB recognition sequences in the promoters of CBF1, CBF2 and CBF3 genes (PubMed:17015446). Involved in drought and salt tolerance. May enhance expression levels of genes involved in abscisic acid (ABA) biosynthesis and signaling, as well as those encoding stress-protective proteins (PubMed:19161942). {ECO:0000269|PubMed:17015446, ECO:0000269|PubMed:19161942}. | |||||
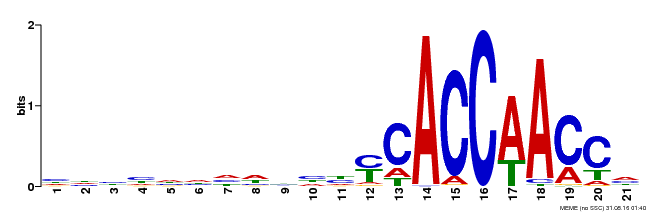
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00375 | DAP | Transfer from AT3G23250 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Aradu.F8RAG |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by abscisic acid (ABA) and drought stress (PubMed:19161942). Induced by salt stress (PubMed:19161942). {ECO:0000269|PubMed:19161942}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF208660 | 0.0 | KF208660.1 Arachis hypogaea putative R2R3 MYB protein 6 (MYB6) gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015940663.1 | 0.0 | myb-related protein Myb4 | ||||
| Refseq | XP_025619598.1 | 0.0 | transcription factor MYB4 | ||||
| Swissprot | Q9LTC4 | 2e-83 | MYB15_ARATH; Transcription factor MYB15 | ||||
| TrEMBL | A0A445BI66 | 0.0 | A0A445BI66_ARAHY; Uncharacterized protein | ||||
| STRING | GLYMA20G35180.1 | 1e-123 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF31 | 34 | 817 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G23250.1 | 4e-81 | myb domain protein 15 | ||||



