 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aradu.L7UKE | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 587aa MW: 66332.4 Da PI: 7.1042 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 86.7 | 3.4e-27 | 89 | 191 | 1 | 91 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkk............tekerrtraetrtgCkaklkvkkekdgkwevtkleleHn 86
+fY+eYA+++GF++ +++s++sk+++e+++++f Cs++g+++e +k+ +e++ +r+ ++t+Cka+++vk+++dgkw+++++++eHn
Aradu.L7UKE 89 SFYQEYARSMGFNTAIQNSRRSKTSREFIDAKFACSRYGTKREYDKSfnrprarqskpdSENSTGRRSCSKTDCKASMHVKRKHDGKWVIHSFVKEHN 186
5********************************************999***99999987777777999****************************** PP
FAR1 87 Helap 91
Hel p
Aradu.L7UKE 187 HELLP 191
**975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 4.5E-25 | 89 | 191 | IPR004330 | FAR1 DNA binding domain |
| PROSITE profile | PS50966 | 9.681 | 308 | 344 | IPR007527 | Zinc finger, SWIM-type |
| Pfam | PF04434 | 1.3E-5 | 315 | 342 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 2.3E-6 | 319 | 346 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 587 aa Download sequence Send to blast |
MDIDLRLPSG EHDKEDEETA TIDNILDGEE KLNNGAMEGR NMVDPGMELQ ALNGGDLNSP 60 TLDMVIFKED TNLEPLSGME FESHGEAYSF YQEYARSMGF NTAIQNSRRS KTSREFIDAK 120 FACSRYGTKR EYDKSFNRPR ARQSKPDSEN STGRRSCSKT DCKASMHVKR KHDGKWVIHS 180 FVKEHNHELL PAQAVSEQTR RMYAAMARQF AEYKTVTSVQ DFVKLYEAIL QDRYEEEAKA 240 DSDTWNKVAT LKTPSPLEKS VAGVYTHAVF KKIQAEVVGA VACHPKADRQ DETTIIHRVH 300 DMQSNKDFFV VVNQVKSEWS CICRLFEYRG YLCRHILIVL QYSGHSAIPS HYILKRWTKD 360 AKVRNIMGEE SEHMLARVQR YNDLCQRAFK LSEEGSLSQE SYSIAVHALH EAQKSCVSVN 420 NSGKSPMEVG TSVTHAQLST EEDTQSRNMG KSNKKKNPAK KKKVNAEAEM LTVGAIDNMQ 480 QMVCPPADKF STRAAVTLDG YYGTQQQVQG MLNLMGPRDD FYGNQQTLQG LGPISSIPTS 540 HDGYYSAHQG MPNLAQLDFL RTGFTYGIRD DPNVRAAQLH EDPSRHA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. When associated with PHYA, protects it from being recognized and degraded by the COP1/SPA complex. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:17012604, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:18715961, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00078 | ChIP-seq | Transfer from AT3G22170 | Download |
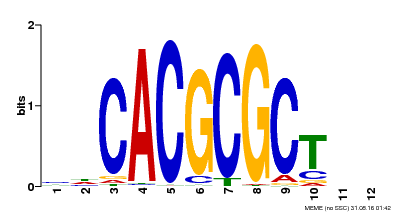
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Aradu.L7UKE |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. Up-regulated by white light. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015040 | 0.0 | AP015040.1 Vigna angularis var. angularis DNA, chromosome 7, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015946937.1 | 0.0 | protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 | ||||
| Refseq | XP_020989242.1 | 0.0 | protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 | ||||
| Refseq | XP_020989243.1 | 0.0 | protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 | ||||
| Refseq | XP_025624745.1 | 0.0 | protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 | ||||
| Refseq | XP_025624747.1 | 0.0 | protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X3 | ||||
| Refseq | XP_029145677.1 | 0.0 | protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 | ||||
| Swissprot | Q9LIE5 | 1e-129 | FHY3_ARATH; Protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| TrEMBL | A0A0A0KV81 | 0.0 | A0A0A0KV81_CUCSA; Uncharacterized protein | ||||
| STRING | XP_007148758.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF40 | 30 | 530 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22170.2 | 1e-127 | far-red elongated hypocotyls 3 | ||||



