 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aradu.M5D2I | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 327aa MW: 35345.3 Da PI: 9.7863 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 50.2 | 5.5e-16 | 257 | 300 | 5 | 49 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkel 49
+r++r++kNRe+A rsR+RK+a+++eLe+kv Le+eN++Lk
Aradu.M5D2I 257 RRQKRMIKNRESAARSRARKQAYTQELENKVSRLEEENERLKR-Q 300
79***************************************94.3 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 1.2E-15 | 249 | 300 | No hit | No description |
| SMART | SM00338 | 4.3E-12 | 253 | 325 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 11.794 | 255 | 300 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 1.1E-12 | 257 | 300 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14707 | 4.18E-24 | 257 | 302 | No hit | No description |
| SuperFamily | SSF57959 | 4.5E-11 | 257 | 302 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 260 | 275 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 327 aa Download sequence Send to blast |
MGSQGGAVQE AQSGGSGSLA RQGSLYNLTL DEVQNQLGNL GKPLGSMNLD ELLKSVFTAE 60 AGGCGGGGGG GQASSGLGSG TDANIQQQHA QLASVSSLNR QGSLTVSGDL SKKTIDEVWR 120 DMQRKKSGGN SNNNDKKTKE TPPTFGEMTL EDFLVKAGVV AESFATEDNA MSRVDSNVAS 180 QHNVSDQVHW IQYQIPSVQP APHLQHNQHQ KQQNNMIAGY IAGPMSPSSL MGTLSDTQTP 240 GRKRVASGTV VEKTVERRQK RMIKNRESAA RSRARKQAYT QELENKVSRL EEENERLKRQ 300 QEIEKVLPSE PPPEPKRQLR RTSSGPL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the embryo specification element and the ABA-responsive element (ABRE) of the Dc3 gene promoter. Could participate in abscisic acid-regulated gene expression during seed development. | |||||
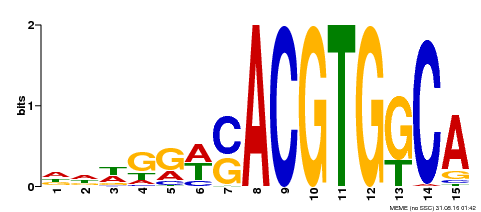
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00409 | DAP | Transfer from AT3G56850 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Aradu.M5D2I |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015970718.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 2 | ||||
| Swissprot | Q9LES3 | 1e-103 | AI5L2_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 2 | ||||
| TrEMBL | A0A445CRV2 | 0.0 | A0A445CRV2_ARAHY; Uncharacterized protein | ||||
| STRING | AES81050 | 1e-127 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1320 | 34 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G56850.1 | 1e-100 | ABA-responsive element binding protein 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



