 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aradu.QNE8S | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 534aa MW: 60458.6 Da PI: 5.5114 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 77.7 | 1.5e-24 | 206 | 259 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k+r++W+ +LH++Fv+av+q+ G +k Pk+il+lm+v+ Lt+e+v+SHLQkYRl
Aradu.QNE8S 206 KARVVWSVDLHQKFVQAVNQI-GFDKVGPKKILDLMNVPWLTRENVASHLQKYRL 259
68*******************.9*******************************8 PP
| |||||||
| 2 | Response_reg | 81.2 | 3.3e-27 | 21 | 129 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTTES CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaGak 95
vl+vdD+p+ +++l+++l+k y +v+++ ++eal++l+e++ +D+++ D++mp+m+G++ll++ e +lp+i+++ ge + + + ++ Ga
Aradu.QNE8S 21 VLVVDDDPTWLKILEKMLKKCSY-QVTTCCLATEALKKLRERKdaFDIVISDVNMPDMNGFKLLEHVGLEM-DLPVIMMSVDGETSRVMKGVQHGAC 115
89*********************.***************888889**********************6644.8************************ PP
EEEESS--HHHHHH CS
Response_reg 96 dflsKpfdpeelvk 109
d+l Kp+ ++el++
Aradu.QNE8S 116 DYLLKPIRMKELRN 129
***********987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 4.4E-144 | 2 | 492 | IPR017053 | Response regulator B-type, plant |
| SuperFamily | SSF52172 | 6.83E-36 | 18 | 143 | IPR011006 | CheY-like superfamily |
| Gene3D | G3DSA:3.40.50.2300 | 3.1E-44 | 18 | 142 | No hit | No description |
| SMART | SM00448 | 2.3E-31 | 19 | 131 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 43.631 | 20 | 135 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 1.4E-24 | 21 | 130 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 2.44E-29 | 22 | 135 | No hit | No description |
| SuperFamily | SSF46689 | 1.97E-17 | 204 | 263 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 5.7E-28 | 204 | 264 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.8E-22 | 206 | 259 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.3E-7 | 208 | 258 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 534 aa Download sequence Send to blast |
MDNNGCLPCT RREGFPAGLR VLVVDDDPTW LKILEKMLKK CSYQVTTCCL ATEALKKLRE 60 RKDAFDIVIS DVNMPDMNGF KLLEHVGLEM DLPVIMMSVD GETSRVMKGV QHGACDYLLK 120 PIRMKELRNI WQHVLRKRIH EGREFESLEG FDFEGIHHLM RNGQDQCCDD VSLFALEDCI 180 SSTRKRKDTD NKHDEREFGD SSSTKKARVV WSVDLHQKFV QAVNQIGFDK VGPKKILDLM 240 NVPWLTRENV ASHLQKYRLY LSRLQKQRDQ KKCSSSGIKN PDLNPKDPGS SSIKNSMNKQ 300 QNVVSIDGFR CPNNGLVQLH ESDFTVSELN KTEQRRALAT NNIIPDPNMT KVSQIVLNQT 360 FSPLNSEGNI EVFDCTIPTQ YTWNQVPKRK LQDDPKSQSI ASMVSSPSAA SSIASSAVDN 420 IIPVQSKSLM VNDQSSVTSN PGLKTQEFDA SLIPDLEFYQ QNLLWGGEAA FSPLEEDLNF 480 FLLQSECYND MNFGMHSIDM SDYYYDPGLI ADVPNHLLYD SADYSVVDQS LFIA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 1e-17 | 202 | 264 | 1 | 63 | ARR10-B |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00010 | PBM | Transfer from AT1G67710 | Download |
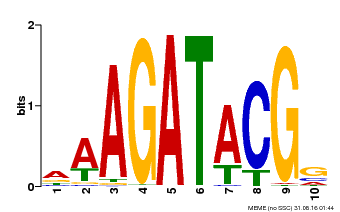
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Aradu.QNE8S |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015957832.1 | 0.0 | two-component response regulator ARR11-like isoform X1 | ||||
| Refseq | XP_020995163.1 | 0.0 | two-component response regulator ARR11-like isoform X1 | ||||
| TrEMBL | A0A445DLL3 | 0.0 | A0A445DLL3_ARAHY; Uncharacterized protein | ||||
| STRING | GLYMA05G27670.2 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3999 | 33 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G67710.1 | 1e-121 | response regulator 11 | ||||



