 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aradu.QV5DJ | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 732aa MW: 78974.8 Da PI: 5.5105 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.7 | 3.3e-16 | 463 | 509 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K++Ka++L +A+eY+k Lq
Aradu.QV5DJ 463 VHNLSERRRRDRINEKMRALQELIPNC-----NKVDKASMLDEAIEYLKTLQ 509
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.60E-17 | 455 | 513 | No hit | No description |
| SuperFamily | SSF47459 | 9.68E-21 | 456 | 520 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 8.9E-21 | 456 | 517 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.349 | 459 | 508 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.1E-13 | 463 | 509 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 7.6E-18 | 465 | 514 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 732 aa Download sequence Send to blast |
MPLYELYRLS REKLDEEING TRATDQSRSS SPEKDFFELI WENGNILTTQ GQSSRAKKSP 60 PRRSLPSHCL PSHSPKGRDR DAGYVNNPRV GKSGDLDTGL NEISMSVPST EVDLGHDDDV 120 IPWLDYTMDG SLQNEYGSNF LHELSGVTDQ DLPSNHFSLV DKSSGNQVFR DSHKNSAEQS 180 NFSSVSSTGV DETTRPKAST VESYLPSSFI SVRPRVSDVT ANNTSNAMLH PPVTEIPSSS 240 SDFSSLKMQK QDQVIPSNGS SVMNFSHFAR PAAIVRANLQ NIGLKSVSSS TRSENVESMN 300 KGAVVPSSNL PESTLADSCS ECPKVLMGNN EKAVEQSRDD LKLLESKSLE QNIAGSKQLD 360 PTCKEKAIKI DQTSNRVLGE TATNTQIAAE RSTELVVASS SVGSGNCADR GSDDPIQNLK 420 RKNRDTEDSE WHSDDVEEES VGVKKAAAGR GASGSKRSRA AEVHNLSERR RRDRINEKMR 480 ALQELIPNCN KVDKASMLDE AIEYLKTLQL QVQMMSMGAG LYMHPMMLPH GMQHMHAPHL 540 APFSPMAYGM QMGLGMGYGM AMPDMNGVSS RFPMVQVPQM QGTHVPVAQM SGPTAMVRSN 600 TQGFGVPGQG FPLPLPRAPL FPFSGGPVMN SSSALGLRPC GTTGLSQTAD LASTSGLKDP 660 SPNTDSLVKQ STGGGGCDST SQMPIQCEAA ATTVGFEQSS MVHNSSHTSE ANDSGTLNPD 720 KEDNNLVTGY DD |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 467 | 472 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
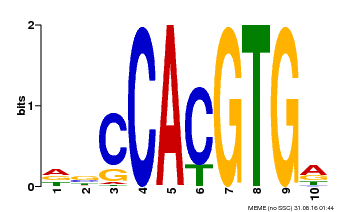
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Aradu.QV5DJ |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015939568.1 | 0.0 | transcription factor PIF3 isoform X2 | ||||
| Refseq | XP_025618802.1 | 0.0 | transcription factor PIF3 isoform X2 | ||||
| TrEMBL | A0A1L2K239 | 0.0 | A0A1L2K239_ARAHY; Phytochrome interacting factor 3 | ||||
| STRING | GLYMA20G22291.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5271 | 33 | 53 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 3e-61 | phytochrome interacting factor 3 | ||||



