 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aradu.RJH1L | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 603aa MW: 68019 Da PI: 6.2097 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 77 | 2.5e-24 | 225 | 278 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k+r++W+ +LH++Fv+av+q+ G +k Pk+il+lm+v+ Lt+e+v+SHLQkYRl
Aradu.RJH1L 225 KARVVWSVDLHQKFVKAVNQI-GFDKVGPKKILDLMNVPWLTRENVASHLQKYRL 278
68*******************.9*******************************8 PP
| |||||||
| 2 | Response_reg | 66.3 | 1.4e-22 | 29 | 153 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH.................ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEEST CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd.................pDlillDiempgmdGlellkeireeepklpiivvtah 80
vl+vdD+p+ +++l+++l+k yee +e + +a ++k + +D+++ D++mp+mdG++ll++ e +lp+i+++
Aradu.RJH1L 29 VLVVDDDPTWLRILEKMLKKCSYEEMNESNSTIKANSWWKVTTcclarhalhllrerkdgYDIVISDVNMPDMDGFKLLEHVGLEM-DLPVIMMSVD 124
89*****************9999999999999999999877777777778888888889***********************6644.8********* PP
TTHHHHHHHHHTTESEEEESS--HHHHHH CS
Response_reg 81 geeedalealkaGakdflsKpfdpeelvk 109
ge + + + ++ Ga d+l Kp+ ++el++
Aradu.RJH1L 125 GETSRVMKGVQHGACDYLLKPIRMKELRN 153
**************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF52172 | 1.57E-29 | 26 | 171 | IPR011006 | CheY-like superfamily |
| Gene3D | G3DSA:3.40.50.2300 | 6.1E-40 | 26 | 190 | No hit | No description |
| SMART | SM00448 | 1.4E-24 | 27 | 155 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 38.206 | 28 | 159 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 1.2E-19 | 29 | 154 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 1.91E-25 | 30 | 159 | No hit | No description |
| SuperFamily | SSF46689 | 4.48E-17 | 223 | 282 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 8.1E-28 | 223 | 283 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.8E-21 | 225 | 278 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.9E-7 | 227 | 277 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 603 aa Download sequence Send to blast |
QRNQPKYFFE LTTRFSSPRR DAFPAGLRVL VVDDDPTWLR ILEKMLKKCS YEEMNESNST 60 IKANSWWKVT TCCLARHALH LLRERKDGYD IVISDVNMPD MDGFKLLEHV GLEMDLPVIM 120 MSVDGETSRV MKGVQHGACD YLLKPIRMKE LRNIWQHVFR KRIHEAREFE SFEGFESIHL 180 MRNGLEQCDD GNLFALEDMT STKKRKDADN KHDDRELIDP SSTKKARVVW SVDLHQKFVK 240 AVNQIGFDKV GPKKILDLMN VPWLTRENVA SHLQKYRLYL SRLQKENDLK SSSSIGMNPD 300 LAATDLGSFV GVQNSVSKKQ NDVSIDSCSY SEGALQLQNM ETKTHEGSSD PNGTISPPAT 360 TEKGRTMIRN FVSEKKMRKN QPFDSLEPEG NHTAVIDCSI PTQFSWNEVP EIQLKDHKPL 420 VQLEGNLSHK FPVTGKKHQI QVDHQSPSIG SIGSPSLTGE EVTACIKTKP LCADYKCEYK 480 SSVSSIGSAV DNNTFPIEPG SLMMNDPSTA PISTTNSGLK TQASNLNSIS DMEFCQRNLL 540 LDAVSLPLDD DLHFNWLHGE CSNINFSLQN IGYYDPGLIA EIPTHYYDSA ADYAVIDQGL 600 FIS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 2e-17 | 221 | 283 | 1 | 63 | ARR10-B |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00010 | PBM | Transfer from AT1G67710 | Download |
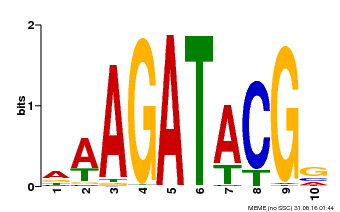
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Aradu.RJH1L |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025622610.1 | 0.0 | two-component response regulator ARR11 isoform X1 | ||||
| TrEMBL | A0A445B9K2 | 0.0 | A0A445B9K2_ARAHY; Two-component response regulator | ||||
| STRING | XP_007163714.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3999 | 33 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G67710.1 | 1e-108 | response regulator 11 | ||||



