 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aradu.V69Z9 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 424aa MW: 48465.3 Da PI: 7.1079 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 59.2 | 8.8e-19 | 126 | 172 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEd +l ++vkq+G++ W+ Ia+++ gR +kqc++rw+++l
Aradu.V69Z9 126 KGQWTPEEDRKLEKLVKQHGTRKWSQIAEKLE-GRAGKQCRERWHNHL 172
799*****************************.*************97 PP
| |||||||
| 2 | Myb_DNA-binding | 53 | 8.2e-17 | 179 | 220 | 2 | 45 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ W++eE+ +lv+ +++ G++ W+ Ia++++ gRt++++k++w+
Aradu.V69Z9 179 DGWSEEEERILVETHAKVGNR-WAEIAKHIP-GRTENSIKNHWN 220
56*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 30.2 | 121 | 176 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.27E-30 | 123 | 219 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.6E-17 | 125 | 174 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-28 | 127 | 180 | IPR009057 | Homeodomain-like |
| Pfam | PF13921 | 4.9E-19 | 129 | 186 | No hit | No description |
| CDD | cd00167 | 2.34E-15 | 129 | 172 | No hit | No description |
| SMART | SM00717 | 6.2E-16 | 177 | 225 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 20.382 | 177 | 227 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-20 | 181 | 225 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 5.07E-12 | 181 | 223 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 424 aa Download sequence Send to blast |
MCRLGLPLTA IDRFLCSQQQ SQVLHNDDSV FDEDFLQTRF SNCYYGGGSN NPYYKLVWPN 60 CTTQEASIVV HHDNNQQEGF NNWLQQIPTL CHNQEDVHQA LGKTNVKVVG RNPKKASSSS 120 SVSLIKGQWT PEEDRKLEKL VKQHGTRKWS QIAEKLEGRA GKQCRERWHN HLRPDIKKDG 180 WSEEEERILV ETHAKVGNRW AEIAKHIPGR TENSIKNHWN ATKRRQNSRR KNRKPSSSTN 240 SNNNGKPQSS ILQDYIRSQT LIGTNNINAP ITTTNSTPTI TSESECDNNN PYYSSPTIIA 300 ESYDDELLFM QQLFKENNQI VNGDGVGKNP SDECGDGFVH YSNNYYYLSN PWNMHNPTTN 360 NNNNGYLDSD LYISQLMNNG VSTTSSSSSL HEAYEKNQNM DFHCSAVKKS EMDLLEFVST 420 HHQF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1mse_C | 9e-43 | 124 | 226 | 2 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 9e-43 | 124 | 226 | 2 | 104 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00567 | DAP | Transfer from AT5G58850 | Download |
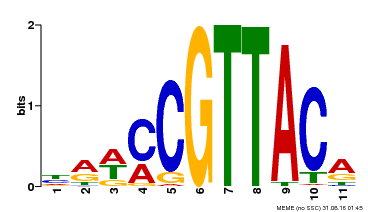
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Aradu.V69Z9 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020985117.1 | 0.0 | transcription factor MYB98-like | ||||
| Refseq | XP_025691244.1 | 0.0 | transcription factor MYB119-like | ||||
| TrEMBL | A0A445EW27 | 0.0 | A0A445EW27_ARAHY; Uncharacterized protein | ||||
| STRING | XP_004500264.1 | 1e-111 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4129 | 31 | 58 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G58850.1 | 9e-61 | myb domain protein 119 | ||||



