 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aradu.VVL06 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 382aa MW: 43659.3 Da PI: 6.9847 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 102.2 | 3.3e-32 | 99 | 152 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtp+LH rFv+av++LGG+e+AtPk +l+lm++kgL+++hvkSHLQ+YR+
Aradu.VVL06 99 PRLRWTPDLHLRFVHAVQRLGGQERATPKLVLQLMNIKGLSIAHVKSHLQMYRS 152
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.929 | 95 | 155 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.4E-29 | 95 | 153 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.31E-14 | 98 | 153 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 6.0E-23 | 99 | 154 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.5E-9 | 100 | 151 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 382 aa Download sequence Send to blast |
MMMMMKMVEE ESHGSECSKT RSPCNDDNNN NEDDGYCEEE SETNNEEEVE EEDESSYKKQ 60 YKNNNGGGGG CSSNSTVEEN NEKKSSSTSV RPYVRSKLPR LRWTPDLHLR FVHAVQRLGG 120 QERATPKLVL QLMNIKGLSI AHVKSHLQMY RSKKVEDTNH QVVMADHHRL LIENGERNNV 180 YNLSQLPMLQ SYRYGHGYGV ASATLGVYEN MAHRNANLTH HHSIFSHNFG EQSSSHPHPP 240 HHHHHHEQKD HQFLSFGNGQ GSLCTNTNNN RLISSHIEKP RAQDFMPTNT NNNLKRKASE 300 NSDIDLDLSL KLNSRVVFND EKQLGRRSSM MMMMMMEEEE HHEVVVNSNL SLSLHSHQSS 360 SPQDHCNKVK GKKATTNLDL RI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 6e-17 | 100 | 154 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 6e-17 | 100 | 154 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 7e-17 | 100 | 154 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 7e-17 | 100 | 154 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 7e-17 | 100 | 154 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 7e-17 | 100 | 154 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 6e-17 | 100 | 154 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 6e-17 | 100 | 154 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 6e-17 | 100 | 154 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 6e-17 | 100 | 154 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 6e-17 | 100 | 154 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 6e-17 | 100 | 154 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00297 | DAP | Transfer from AT2G38300 | Download |
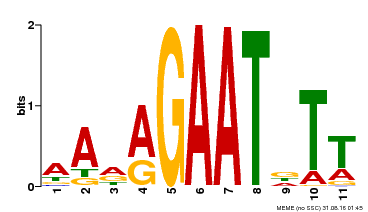
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Aradu.VVL06 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015961024.1 | 0.0 | transcription factor LUX-like isoform X1 | ||||
| TrEMBL | A0A445DFA5 | 0.0 | A0A445DFA5_ARAHY; Uncharacterized protein | ||||
| STRING | AES79549 | 6e-79 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2378 | 34 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38300.1 | 9e-46 | G2-like family protein | ||||



