 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aradu.W58GD | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 354aa MW: 37490.4 Da PI: 9.6217 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 121.9 | 2.2e-38 | 75 | 134 | 3 | 62 |
zf-Dof 3 ekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
e a+kcprC+stntkfCy+nnyslsqPr+fCk+CrryWt+GGalrnvPvGgg+r+nk+++
Aradu.W58GD 75 EPAQKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGCRRNKRTK 134
56899****************************************************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 2.0E-29 | 58 | 123 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 3.8E-32 | 77 | 132 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 29.311 | 78 | 132 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 80 | 116 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 354 aa Download sequence Send to blast |
MVYTSIPAYI DPVNWHQQQQ PNHQQNNTGG TSSQLLHPPP PPLPPSTQQH GASTSSIRPG 60 SMADRARMAN IPMQEPAQKC PRCESTNTKF CYFNNYSLSQ PRHFCKTCRR YWTRGGALRN 120 VPVGGGCRRN KRTKGTTTSN NGSKSPPPSS DRGGPTVSGG SFASEIAGLS PPVPSSLRFM 180 APLMHHQQLG DHFSGVGGGD LGLNYAAISG PMGGMGDLNF HIGSALRSDV TGGGNGGSSL 240 LSAAGLEQWR VPQTQQFSFL TGFEASSHGL YQFENSTSEG SGYGGGSSGG GGAIRPNKVS 300 TFASVVKMEE SNNNLPRQFL GMNNNSSSNS EHFVPASWTH LSGFTSSNTT NNPP |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00295 | DAP | Transfer from AT2G37590 | Download |
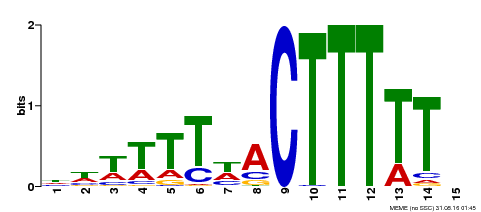
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Aradu.W58GD |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015960388.1 | 0.0 | dof zinc finger protein DOF2.4-like isoform X1 | ||||
| Refseq | XP_025695227.1 | 0.0 | dof zinc finger protein DOF2.4 isoform X1 | ||||
| TrEMBL | A0A444WQI6 | 0.0 | A0A444WQI6_ARAHY; Uncharacterized protein | ||||
| STRING | XP_004515618.1 | 1e-105 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF656 | 34 | 143 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G55370.1 | 5e-39 | OBF-binding protein 3 | ||||



