 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aradu.Z2BLJ | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 338aa MW: 37790.6 Da PI: 4.4823 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 58.7 | 9.6e-19 | 61 | 114 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ eq+++Le+ Fe ++++ +++ +LA++lgL+ rqV vWFqNrRa++k
Aradu.Z2BLJ 61 KKRRLSVEQVKALEKNFEVENKLEPDRKVKLAQELGLQPRQVAVWFQNRRARWK 114
456899***********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 130.3 | 7.6e-42 | 60 | 151 | 1 | 92 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelk 92
ekkrrls eqvk+LE++Fe e+kLep+rKv+la+eLglqprqvavWFqnrRAR+ktkqlE+dy +Lk++y+alk + e+L++++++L++e+k
Aradu.Z2BLJ 60 EKKRRLSVEQVKALEKNFEVENKLEPDRKVKLAQELGLQPRQVAVWFQNRRARWKTKQLERDYGVLKANYEALKLNFEKLQQDNQALHKEIK 151
69**************************************************************************************9997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 9.84E-20 | 44 | 118 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.378 | 56 | 116 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.7E-18 | 59 | 120 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.44E-16 | 61 | 117 | No hit | No description |
| Pfam | PF00046 | 7.2E-16 | 61 | 114 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 4.5E-20 | 63 | 123 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 9.9E-6 | 87 | 96 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 91 | 114 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 9.9E-6 | 96 | 112 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 1.3E-17 | 116 | 158 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 338 aa Download sequence Send to blast |
MKRLSSSDDS LGALMTICPS STTEEHSPRN KQQVYGREFQ SMLDGLDEDG CIEESGHHGE 60 KKRRLSVEQV KALEKNFEVE NKLEPDRKVK LAQELGLQPR QVAVWFQNRR ARWKTKQLER 120 DYGVLKANYE ALKLNFEKLQ QDNQALHKEI KGLKSRMQDE RTESDASVKK EQVEMITTLQ 180 DSEENPCDLE TAAAIAGSDS NKDLSYDCFN KGDGEGGDGV GVAASISLFP CGDLKDGSSD 240 SDSSAILNEE NNNSSCSPMN NAAMSSCGGG VLQSSNLLMS QHDESPSSMS CFQFQKAYQA 300 TQYVKMEEHN FFSADETCNF FSDEQAPTLQ WYCSEEWS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 108 | 116 | RRARWKTKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00271 | DAP | Transfer from AT2G22430 | Download |
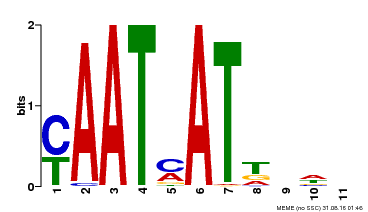
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Aradu.Z2BLJ |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015957630.1 | 0.0 | homeobox-leucine zipper protein ATHB-6 isoform X1 | ||||
| TrEMBL | A0A444ZTV8 | 0.0 | A0A444ZTV8_ARAHY; Uncharacterized protein | ||||
| STRING | XP_004504111.1 | 1e-133 | (Cicer arietinum) | ||||
| STRING | GLYMA05G30940.1 | 1e-133 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1868 | 34 | 89 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22430.1 | 2e-75 | homeobox protein 6 | ||||



