 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.0408s0004.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 414aa MW: 46906.4 Da PI: 7.5848 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 11.3 | 0.0011 | 35 | 57 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
y C++Cg s s k++ ++Hi++H
Araha.0408s0004.1.p 35 YLCQYCGISRSKKYLITSHIQSH 57
56********************6 PP
| |||||||
| 2 | zf-C2H2 | 20.1 | 1.7e-06 | 81 | 103 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
++C+ Cg F+ + +Lk+H+++H
Araha.0408s0004.1.p 81 HTCHECGAEFKKPAHLKQHMQSH 103
79*******************99 PP
| |||||||
| 3 | zf-C2H2 | 16 | 3.5e-05 | 109 | 133 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ C dC +s++rk++L+rH+ tH
Araha.0408s0004.1.p 109 FACYvdDCATSYRRKDHLNRHLLTH 133
668888*****************99 PP
| |||||||
| 4 | zf-C2H2 | 22.2 | 3.7e-07 | 217 | 240 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirtH 23
+C+ Cgk F+ +s+L++H+ +H
Araha.0408s0004.1.p 217 VCKeiGCGKAFKYPSQLQKHQDSH 240
79999****************998 PP
| |||||||
| 5 | zf-C2H2 | 19.6 | 2.4e-06 | 306 | 331 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
+kC+ C+ +Fs snL++H+r H
Araha.0408s0004.1.p 306 FKCEveGCSSTFSKASNLQKHLRAvH 331
89*********************888 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00355 | 18 | 35 | 57 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 13.858 | 81 | 108 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 4.3E-4 | 81 | 100 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0044 | 81 | 103 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 83 | 103 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.23E-10 | 94 | 135 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.1E-12 | 101 | 135 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 9.889 | 109 | 138 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.34 | 109 | 133 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 111 | 133 | IPR007087 | Zinc finger, C2H2 |
| PROSITE profile | PS50157 | 9.307 | 138 | 168 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.3 | 138 | 163 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 140 | 163 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.2E-6 | 211 | 240 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 1.82E-5 | 211 | 242 | No hit | No description |
| PROSITE profile | PS50157 | 10.72 | 216 | 245 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0028 | 216 | 240 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 218 | 240 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.97 | 248 | 273 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 8.9E-4 | 250 | 271 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 250 | 273 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 22 | 276 | 297 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 2.3E-8 | 287 | 328 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 2.33E-9 | 287 | 344 | No hit | No description |
| PROSITE profile | PS50157 | 13.401 | 306 | 336 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 3.2E-4 | 306 | 331 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 308 | 331 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 8.6E-5 | 330 | 357 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 8.1 | 337 | 363 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 339 | 363 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 414 aa Download sequence Send to blast |
MEEINPSSCV DNQRDMAEEA KKDLKTSAKK DIRNYLCQYC GISRSKKYLI TSHIQSHHQM 60 ELEEEKDDET CEVEEESSSK HTCHECGAEF KKPAHLKQHM QSHSLERPFA CYVDDCATSY 120 RRKDHLNRHL LTHKGKLFKC PKENCKSEFS VQGNVSRHVK KCHSSGDSDK DTTGKGDSDK 180 EKSGDKENTG NGDSGKENTG NGDSQPSECS TGQKKLVCKE IGCGKAFKYP SQLQKHQDSH 240 VKLDSVEAFC SEPGCMKYFT NEECLKAHIR SCHQHINCDI CGSKHLKKNI KRHLRTHDED 300 SSPGEFKCEV EGCSSTFSKA SNLQKHLRAV HEDLRPFVCG FPGCGMRFAY KHVRNNHENS 360 GCHVYTCGDF VETDEDFTSR PRGGLKRKQV TAEMLVRKRV MPPRFDSEEH ETC* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wjq_D | 2e-13 | 33 | 357 | 7 | 275 | Zinc finger protein 568 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
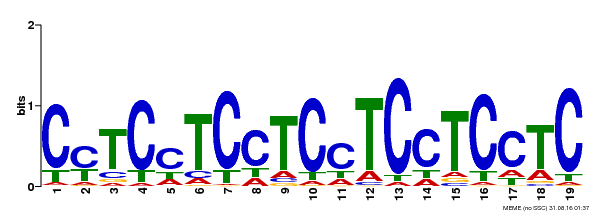
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.0408s0004.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AJ630478 | 0.0 | AJ630478.1 Arabidopsis thaliana mRNA for hypothetical protein, clone At1g72050D. | |||
| GenBank | AY054225 | 0.0 | AY054225.1 Arabidopsis thaliana At1g72050/F28P5_6 mRNA, complete cds. | |||
| GenBank | AY066042 | 0.0 | AY066042.1 Arabidopsis thaliana At1g72050/F28P5_6 mRNA, complete cds. | |||
| GenBank | AY186610 | 0.0 | AY186610.1 Arabidopsis thaliana transcription factor IIIA (TFIIIA) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020891257.1 | 0.0 | transcription factor IIIA | ||||
| Swissprot | Q84MZ4 | 0.0 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A1P8AV27 | 0.0 | A0A1P8AV27_ARATH; Transcription factor IIIA | ||||
| STRING | Bostr.10273s0148.1.p | 0.0 | (Boechera stricta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4714 | 28 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 0.0 | transcription factor IIIA | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.0408s0004.1.p |



