 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.10347s0003.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 464aa MW: 51235.9 Da PI: 8.1646 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 96.2 | 2.2e-30 | 110 | 168 | 1 | 60 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
++DgynWrKYGqK vkg+ef rsYYrCt+++C++kk++ers+ ++vv+++Y geH+h+k
Araha.10347s0003.1.p 110 MEDGYNWRKYGQKLVKGNEFVRSYYRCTHPNCKAKKQLERSS-GGQVVDTVYFGEHDHPK 168
58****************************************.***************85 PP
| |||||||
| 2 | WRKY | 105.8 | 2.3e-33 | 282 | 340 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
++Dgy+WrKYGqK+vkgs++prsYYrC+s+gCpvkk+vers++d+k++++tYeg+H+h+
Araha.10347s0003.1.p 282 VNDGYRWRKYGQKSVKGSPYPRSYYRCSSSGCPVKKHVERSSHDTKLLITTYEGKHDHD 340
58********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50811 | 22.745 | 105 | 169 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 9.15E-25 | 106 | 169 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 2.1E-26 | 107 | 169 | IPR003657 | WRKY domain |
| SMART | SM00774 | 6.9E-35 | 110 | 168 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 7.1E-24 | 111 | 167 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.2E-34 | 268 | 342 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.31E-28 | 276 | 342 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 34.991 | 277 | 342 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.3E-38 | 282 | 341 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.9E-26 | 283 | 339 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009863 | Biological Process | salicylic acid mediated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 464 aa Download sequence Send to blast |
MAEVGKVPAS DMELDHSSET KAGDDVVSAT DKAEVTPVTV TITETVVESS KSTDCKELQQ 60 LVPHTVALQS EVDVASPVSE KAPKISESST ALSLQSGSEG NSPFIREKVM EDGYNWRKYG 120 QKLVKGNEFV RSYYRCTHPN CKAKKQLERS SGGQVVDTVY FGEHDHPKPL TGAVLINQDK 180 RSDVFTAVSK EKSSGSSVQA HRQTEPPKIH GGLHVSVIPP ADDVKTDISQ SSRIKGDSTH 240 KDHTSPSSKR RKKGGNIELS PVERSTNDSR IVVHTQTLFD IVNDGYRWRK YGQKSVKGSP 300 YPRSYYRCSS SGCPVKKHVE RSSHDTKLLI TTYEGKHDHD MPPGRVVTHN NMLDSEVDDK 360 EGDANKTPQS SALQSITKDQ RVEDHPRKKT KTNGFEKSLD QGPVLDEKLK EKIKERSDVN 420 KDQAANHAKP ETKSDDKTTA CHEKAVVTLE NEEQKPKTEP ARS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 4e-49 | 109 | 344 | 1 | 76 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
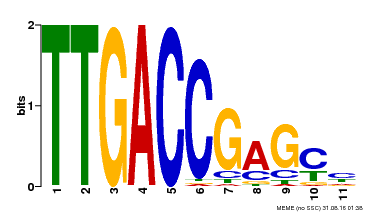
| UniProt | Transcription factor. Binds to a 5'-CGTTGACCGAG-3' consensus core sequence which contains a W box, a frequently occurring elicitor-responsive cis-acting element. {ECO:0000269|PubMed:8972846}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00094 | SELEX | Transfer from AT2G04880 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.10347s0003.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salicylic acid (SA). {ECO:0000269|PubMed:17264121}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF442390 | 0.0 | AF442390.1 Arabidopsis thaliana WRKY transcription factor 1 splice variant 2 (WRKY1) mRNA, complete cds; alternatively spliced. | |||
| GenBank | BT029167 | 0.0 | BT029167.1 Arabidopsis thaliana At2g04880 mRNA, complete cds. | |||
| GenBank | X92976 | 0.0 | X92976.1 A.thaliana mRNA for ZAP1. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020886221.1 | 0.0 | WRKY transcription factor 1 | ||||
| Swissprot | Q9SI37 | 0.0 | WRKY1_ARATH; WRKY transcription factor 1 | ||||
| TrEMBL | C0SV43 | 0.0 | C0SV43_ARATH; Uncharacterized protein At2g04880 (Fragment) | ||||
| STRING | Al_scaffold_0003_2954 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM7949 | 26 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G04880.2 | 0.0 | zinc-dependent activator protein-1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.10347s0003.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



