 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.10517s0003.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 818aa MW: 91515.7 Da PI: 5.396 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 135 | 2.4e-42 | 121 | 197 | 2 | 78 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
Cqve+C adls++k+yhrrhkvCe+hska+++lv g++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
Araha.10517s0003.1.p 121 CQVENCGADLSKVKDYHRRHKVCEMHSKATSALVGGIMQRFCQQCSRFHVLEEFDEGKRSCRRRLAGHNKRRRKANP 197
**************************************************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.1E-34 | 116 | 182 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.565 | 118 | 195 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.31E-39 | 120 | 199 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 3.6E-31 | 121 | 194 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 1.7E-6 | 706 | 810 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 9.24E-7 | 708 | 809 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 5.11E-8 | 711 | 809 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 818 aa Download sequence Send to blast |
MEARIEGEVE GHSLGYGFSG KRSVEWDLND WKWNGDLFVA TQLNHGSSNS SSTCSDEANV 60 EIMEKKKKRG AITVIAVEED NLRDGDDDAH RLTLNLGGNI EGNGAKKTKL GGGIPSRAIS 120 CQVENCGADL SKVKDYHRRH KVCEMHSKAT SALVGGIMQR FCQQCSRFHV LEEFDEGKRS 180 CRRRLAGHNK RRRKANPDIG NGNPLSDDQT SNYMLITLLK ILSNIHTNQS DQTGDQDLLS 240 HLLKSLVSQA GEHIGRNLVG LLQGGVGLQA SQNIGNSSAL LSLEQAPRED IKHHSVPETN 300 WQEVYANGAQ ETAAPDRSEK QVKMNDFDLN DIYIDSDDTT DIERSSPPPT NPATSSLDYH 360 QDSRQSSPPQ TSRRNSDSAS DQSPSSSSGD GQSRTDRIVF KLFGKEPNDF PVALRGQILN 420 WLAHTPTDME SYIRPGCIVL TIYLRQDEAS WEELCCDLSF SLRRLLDLSD DPLWTDGWIY 480 LRVQNQLAFA FDGQVVLDTS LPLRSHDYSQ IIAVRPLAVT RKAQFTVKGI NLRRPGTRLL 540 CAVEGTYLVQ EATQGVMEER DELKENNEID FVKFSCEMPI ASGRGFMEIE DQGGLSSSYF 600 PFIVSEDEDV CSEIRRLEST LEFTGTDSAM QAMDFIHEIG WLLHRSELKS RLAASDHNPE 660 DLFPLIRFKF LIEFSMDREW YAVIKKLLNI LFEEGTVDPS PDAALSELCL LHRAVRKNSK 720 PMVEMLLRFI PKKMNQTLSG LFRPDAAGPG GLTPLHIAAG KDGSEDVLDA LTEDPDMIGI 780 QAWKISRDNT GFTPEDYARL RGHFSYIHLV QRKLNRKP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj0_A | 4e-34 | 118 | 175 | 3 | 60 | squamosa promoter-binding protein-like 12 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
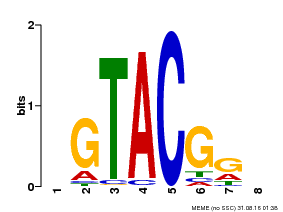
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000269|PubMed:16554053}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00060 | PBM | Transfer from AT3G60030 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.10517s0003.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB493658 | 0.0 | AB493658.1 Arabidopsis thaliana At3g60030 mRNA for hypothetical protein, partial cds, clone: RAAt3g60030. | |||
| GenBank | AJ132096 | 0.0 | AJ132096.1 Arabidopsis thaliana mRNA for squamosa promoter binding protein-like 12. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002878311.1 | 0.0 | squamosa promoter-binding-like protein 12 | ||||
| Refseq | XP_020880464.1 | 0.0 | squamosa promoter-binding-like protein 12 | ||||
| Refseq | XP_020880465.1 | 0.0 | squamosa promoter-binding-like protein 12 | ||||
| Swissprot | Q9S7P5 | 0.0 | SPL12_ARATH; Squamosa promoter-binding-like protein 12 | ||||
| TrEMBL | D7LWK1 | 0.0 | D7LWK1_ARALL; Uncharacterized protein | ||||
| STRING | scaffold_503227.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3030 | 27 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G60030.1 | 0.0 | squamosa promoter-binding protein-like 12 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.10517s0003.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



