 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.10571s0003.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 250aa MW: 27554.1 Da PI: 9.5553 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 98.8 | 3.5e-31 | 171 | 228 | 3 | 59 |
-SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 3 DgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
D+y+WrKYGqK++kgs++pr+YY+C++ gCp++k+ver+ +dp ++++tYegeH+h+
Araha.10571s0003.1.p 171 DEYSWRKYGQKPIKGSPHPRGYYKCSTFrGCPARKHVERALDDPAMLIVTYEGEHRHN 228
9************************8766****************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF10533 | 7.8E-18 | 118 | 167 | IPR018872 | Zn-cluster domain |
| Gene3D | G3DSA:2.20.25.80 | 5.0E-33 | 157 | 229 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 30.24 | 164 | 230 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 9.81E-27 | 167 | 229 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.8E-37 | 169 | 229 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 4.9E-27 | 171 | 227 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:1990841 | Molecular Function | promoter-specific chromatin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 250 aa Download sequence Send to blast |
MAVDLMRFPK MDDQTAIQEA ASQGLQSMEH LIRVLSNRPE QQHNNVDCSE ITDFTVSKFK 60 TLEFSKENFS VSLNSSFMSS GITGDGSVSN GKIFLASAPS QPVNSSGKPP LAAGHPYRKR 120 CLEHEHSESF SGKVSGSGYG KCHCKKSRKN RMKRTVRVPA ISAKIADIPP DEYSWRKYGQ 180 KPIKGSPHPR GYYKCSTFRG CPARKHVERA LDDPAMLIVT YEGEHRHNQS AMQENISSSG 240 VNDLVFASA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 9e-21 | 157 | 228 | 2 | 72 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). Regulates rhizobacterium B.cereus AR156-induced systemic resistance (ISR) to P.syringae pv. tomato DC3000, probably by activating the jasmonic acid (JA)- signaling pathway (PubMed:26433201). {ECO:0000250, ECO:0000269|PubMed:26433201}. | |||||
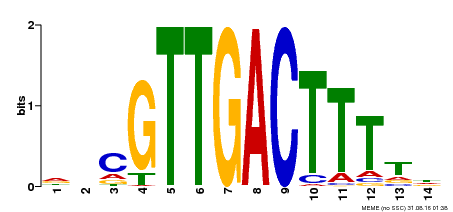
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00464 | DAP | Transfer from AT4G31550 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.10571s0003.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Strongly during leaf senescence (PubMed:11722756). Repressed by rhizobacterium B.cereus AR156 in leaves, and to a lower extent, by P.fluorescens WCS417r (PubMed:26433201). {ECO:0000269|PubMed:11722756, ECO:0000269|PubMed:26433201}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY150394 | 0.0 | AY150394.1 Arabidopsis thaliana putative DNA-binding protein (At4g31550) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002867294.1 | 1e-139 | probable WRKY transcription factor 11 isoform X1 | ||||
| Swissprot | Q9SV15 | 1e-137 | WRK11_ARATH; Probable WRKY transcription factor 11 | ||||
| TrEMBL | A0A078J443 | 1e-138 | A0A078J443_BRANA; BnaA01g34790D protein | ||||
| TrEMBL | D7MB53 | 1e-138 | D7MB53_ARALL; WRKY transcription factor 11 | ||||
| STRING | scaffold_701056.1 | 1e-138 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2508 | 27 | 72 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G31550.1 | 1e-127 | WRKY DNA-binding protein 11 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.10571s0003.1.p |



