 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.10968s0006.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 350aa MW: 37731.6 Da PI: 7.2021 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 125.3 | 2e-39 | 44 | 105 | 2 | 63 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkksss 63
k++al+cprC+s ntkfCyynnysl+qPryfCk CrryWt+GG+lrn+PvGgg rknk+sss
Araha.10968s0006.1.p 44 KDQALNCPRCNSLNTKFCYYNNYSLTQPRYFCKDCRRYWTAGGSLRNIPVGGGVRKNKRSSS 105
7899******************************************************9875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 5.0E-33 | 30 | 96 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 2.7E-33 | 45 | 102 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 28.863 | 48 | 102 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 50 | 86 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 350 aa Download sequence Send to blast |
MDTAKWPQEF VVKPMNEIVT NTCLKQPNHP PTATPVERKA RPEKDQALNC PRCNSLNTKF 60 CYYNNYSLTQ PRYFCKDCRR YWTAGGSLRN IPVGGGVRKN KRSSSSSPSS SSSSKKPLFA 120 NNNTPTPPLP QRNPEIGEAA ATKVQDLTFS PGFGNAHEVK DLNLAFSQGF GIAHHHHSHS 180 SIPEFLQVVP SSSMKNNTLV STSSALELLG ISTSSNSSRP AFMSYPNVHD SSVYTASGFG 240 LSYPQFQDFM RPALGFSLDG GDPLHQEEGS SGTNNGRPLL PFESLKLPVS SSSTNSGGND 300 NVKEINDHHS DHDEHEKEEG EADHSVGFWN GMLSAGASAA AASGGGSWQ* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00210 | DAP | Transfer from AT1G64620 | Download |
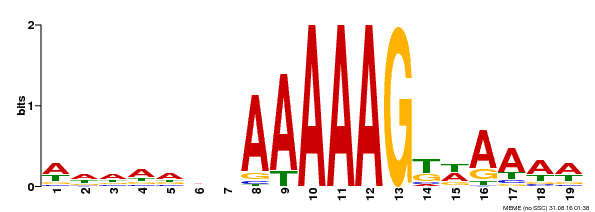
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.10968s0006.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY084898 | 0.0 | AY084898.1 Arabidopsis thaliana clone 120488 mRNA, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002887870.1 | 0.0 | dof zinc finger protein DOF1.8 | ||||
| Swissprot | Q84JQ8 | 0.0 | DOF18_ARATH; Dof zinc finger protein DOF1.8 | ||||
| TrEMBL | D7KS75 | 0.0 | D7KS75_ARALL; Uncharacterized protein | ||||
| STRING | fgenesh2_kg.2__13__AT1G64620.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM14932 | 17 | 20 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G64620.1 | 1e-170 | Dof family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.10968s0006.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



