 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.11451s0009.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 548aa MW: 63117.4 Da PI: 8.6344 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 95.6 | 4.7e-30 | 12 | 96 | 1 | 87 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqlea 87
rW++ e+laL+++r+em++++r++ lk+plWee+s+km+e g++rs+k+Ckek+en+ k++k++keg+ ++++++ t+++f++lea
Araha.11451s0009.1.p 12 RWPRPETLALLRIRSEMDKAFRDSTLKAPLWEEISRKMMELGYKRSSKKCKEKFENVYKYHKRTKEGRTGKSEGK--TYRFFEELEA 96
8********************************************************************975544..6******985 PP
| |||||||
| 2 | trihelix | 108.9 | 3.2e-34 | 368 | 453 | 1 | 87 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqlea 87
rW+k ev aLi++r+++e +++++ k+plWee+s+ mr+ g++rs+k+Ckekwen+nk++kk+ke++kkr + +s+tcpyf+qlea
Araha.11451s0009.1.p 368 RWPKTEVEALIRIRKNLEANYQENGTKGPLWEEISAGMRRLGYNRSAKRCKEKWENINKYFKKVKESNKKR-PLDSKTCPYFHQLEA 453
8*********************************************************************8.9************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 0.086 | 9 | 71 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 4.10E-24 | 11 | 76 | No hit | No description |
| Pfam | PF13837 | 4.8E-20 | 11 | 96 | No hit | No description |
| PROSITE profile | PS50090 | 7.073 | 11 | 69 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 9.2E-4 | 365 | 427 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 6.6E-24 | 367 | 454 | No hit | No description |
| CDD | cd12203 | 1.41E-25 | 367 | 432 | No hit | No description |
| PROSITE profile | PS50090 | 7.491 | 367 | 425 | IPR017877 | Myb-like domain |
| Gene3D | G3DSA:1.10.10.60 | 3.1E-4 | 367 | 424 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 4.44E-5 | 368 | 459 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 548 aa Download sequence Send to blast |
ETGEGAGGGG NRWPRPETLA LLRIRSEMDK AFRDSTLKAP LWEEISRKMM ELGYKRSSKK 60 CKEKFENVYK YHKRTKEGRT GKSEGKTYRF FEELEAFETL SSYQPEPESQ PAKSSAAVTT 120 APATTSLIPW ISSSNPSIEK ISSPLKHHHQ VSAQPITTNP TFLAKQPSST TPFPFYSNNN 180 TTTVSQPPIS NDLMNNVSSL NLFSNSTSSS TASDEEEDHH QGKRSRKKRK YWKGLFTKLT 240 KELMEKQKKM QKRFLETLEN REKERITREE AWRVQEIARI NREHEILIHE RSNAAAKDAA 300 IISFLHKISG GQPQQPQQHN HKPSQRKQYR SDHSITFESK EPRPVLLDTT MKMGNYDNNH 360 LVSPSSSRWP KTEVEALIRI RKNLEANYQE NGTKGPLWEE ISAGMRRLGY NRSAKRCKEK 420 WENINKYFKK VKESNKKRPL DSKTCPYFHQ LEALYNERNK SGTMPLPLPL MVTPQRQLLL 480 SQETQTESET DQRDKVGDKE DEEEEGESEE DEYDEEEEGE GDNETSEFEI VLNKTSSPMD 540 INNNLFT* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 54 | 62 | KRSSKKCKE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds specific DNA sequence. {ECO:0000250}. | |||||
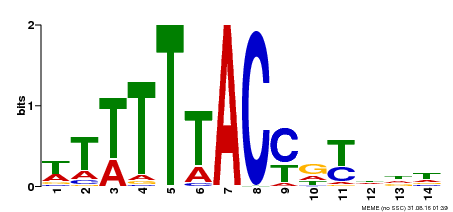
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00244 | DAP | Transfer from AT1G76890 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.11451s0009.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY087117 | 0.0 | AY087117.1 Arabidopsis thaliana clone 3190 mRNA, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002889107.1 | 0.0 | trihelix transcription factor GT-2 | ||||
| Swissprot | Q39117 | 0.0 | TGT2_ARATH; Trihelix transcription factor GT-2 | ||||
| TrEMBL | D7KTY0 | 0.0 | D7KTY0_ARALL; Uncharacterized protein | ||||
| STRING | scaffold_202644.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5995 | 28 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G76890.2 | 0.0 | Trihelix family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.11451s0009.1.p |



