 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.1146s0009.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 480aa MW: 54592 Da PI: 6.3376 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 122.4 | 3.9e-38 | 6 | 137 | 3 | 128 |
NAM 3 pGfrFhPtdeelvveyLkkkvegkk.leleevikevdiykveP.wdLpk..kvkaeekewyfFskrdkkyatgkrknratksgyWkatgk 88
G+rF+Pt eel+++yLk+k g+k l ++e+i+e++i++++P +dLpk k+++e+ ewyfFs+ + +++++++ +r+t sg+Wk tg
Araha.1146s0009.1.p 6 IGYRFSPTGEELINHYLKNKNLGTKtLLVDEAISEINILSHKPsKDLPKlaKIQSEDLEWYFFSPIEYTNRKKNEMKRTTGSGFWKPTGV 95
69******************888874666678***********558995448888888******************************** PP
NAM 89 dkevlsk..kgelvglkktLvfykgrapkgektdWvmheyrl 128
d+++++k +g ++g kktLv+++g+ p+g++t W mhey++
Araha.1146s0009.1.p 96 DRKIKDKrgNGVVIGFKKTLVYHEGKGPNGVRTPWAMHEYHI 137
******9777788***************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51005 | 44.317 | 4 | 155 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 1.15E-43 | 5 | 155 | IPR003441 | NAC domain |
| Pfam | PF02365 | 6.8E-22 | 7 | 137 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009736 | Biological Process | cytokinin-activated signaling pathway | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051302 | Biological Process | regulation of cell division | ||||
| GO:0005789 | Cellular Component | endoplasmic reticulum membrane | ||||
| GO:0031965 | Cellular Component | nuclear membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 480 aa Download sequence Send to blast |
MVKDLIGYRF SPTGEELINH YLKNKNLGTK TLLVDEAISE INILSHKPSK DLPKLAKIQS 60 EDLEWYFFSP IEYTNRKKNE MKRTTGSGFW KPTGVDRKIK DKRGNGVVIG FKKTLVYHEG 120 KGPNGVRTPW AMHEYHITSL PHDKRKYVIC QVKYKGEAAD ISYEPSHSLD SDSNTTRAAE 180 PRLQVEQPGR ENFLGMSLDD LEQEDPNTYL QPQGPHLALN DDEFIRGLRH VDRGPVEYLF 240 ANEENVDGLS MNDLRIPMIV QQEDLSEWEG FNADTFFSDN NNTHNLNVHQ LTRYGDDYQN 300 AGYNGGNFEG VHPDQELIMQ ENRNDHMPKK PLTGTIDYSS DSGSDAGSIS TTSYQGISSP 360 NISVGSSSRR LSSFSSTDSC KDLQTSADPS ISRETREYQL TQRTVPSKQE VKQEIPRAVD 420 ASMNNESSLV KTEEKCLFIL EDAMERNRKT PRFIYLMIIG FIRRIGFTTR QRLNPLKKF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 2e-22 | 7 | 137 | 18 | 140 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator activated by proteolytic cleavage through regulated intramembrane proteolysis (RIP) mediated by calpain or its functional homolog. Regulates cytokinin signaling during cell division. {ECO:0000269|PubMed:17098812}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00425 | DAP | Transfer from AT4G01540 | Download |
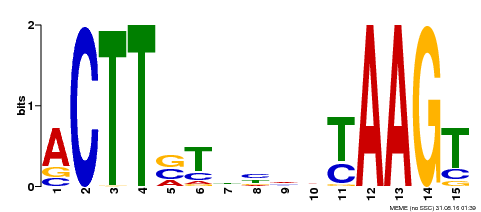
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.1146s0009.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Stabilized by cytokinins. {ECO:0000269|PubMed:17098812}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB493671 | 0.0 | AB493671.1 Arabidopsis thaliana At4g01540 mRNA for hypothetical protein, partial cds, clone: RAAt4g01540. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002872887.1 | 0.0 | NAC domain-containing protein 68 | ||||
| Swissprot | A8MQY1 | 0.0 | NAC68_ARATH; NAC domain-containing protein 68 | ||||
| TrEMBL | D7M4U2 | 0.0 | D7M4U2_ARALL; ANAC068/NTM1 | ||||
| STRING | scaffold_604133.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1982 | 19 | 71 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G01540.1 | 0.0 | NAC with transmembrane motif1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.1146s0009.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



