 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.13692s0022.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1030aa MW: 115773 Da PI: 5.9826 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 186 | 3.9e-58 | 21 | 138 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahse 89
l+e ++rwl+++ei++iL+n++k+++++e+++rp sgsl+L++rk++ryfrkDG++w+kkkdgkt++E+hekLKvg+++vl+cyYah+e
Araha.13692s0022.1.p 21 LSEaQHRWLRPSEICEILQNYHKFHIASESPSRPASGSLFLFDRKVLRYFRKDGHNWRKKKDGKTIKEAHEKLKVGSIDVLHCYYAHGE 109
45559************************************************************************************ PP
CG-1 90 enptfqrrcywlLeeelekivlvhylevk 118
n++fqrrcyw+Le++l +iv+vhylevk
Araha.13692s0022.1.p 110 GNENFQRRCYWMLEQDLMHIVFVHYLEVK 138
**************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 84.235 | 17 | 143 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.4E-83 | 20 | 138 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 7.8E-51 | 23 | 137 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 2.1E-14 | 434 | 520 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 5.2E-4 | 434 | 509 | IPR002909 | IPT domain |
| Gene3D | G3DSA:2.60.40.10 | 3.0E-4 | 435 | 509 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF12796 | 2.1E-7 | 616 | 694 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 19.81 | 617 | 728 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 7.6E-18 | 618 | 729 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 5.75E-19 | 618 | 728 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.04E-14 | 623 | 726 | No hit | No description |
| SMART | SM00248 | 3700 | 634 | 663 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 8.603 | 634 | 666 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.22 | 667 | 699 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.0029 | 667 | 696 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 0.51 | 842 | 864 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.73 | 843 | 872 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 1.04E-7 | 843 | 892 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| Pfam | PF00612 | 0.0025 | 845 | 863 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0092 | 865 | 887 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.395 | 866 | 890 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.5E-4 | 868 | 887 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050826 | Biological Process | response to freezing | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1030 aa Download sequence Send to blast |
MVDRGSFGFI SPPQLDMEQL LSEAQHRWLR PSEICEILQN YHKFHIASES PSRPASGSLF 60 LFDRKVLRYF RKDGHNWRKK KDGKTIKEAH EKLKVGSIDV LHCYYAHGEG NENFQRRCYW 120 MLEQDLMHIV FVHYLEVKGN RTSIGMKENN SNSVNGTASV NIDSTASPTS TLSSLCEDAD 180 TGDSHQASSV LRASSEPQTG NRYGWTPAPG MRNVSQVHGN RVRESDSQRL VDVRAWDAVG 240 NSVTRYHGQP YCNNLLTPMQ PSNTDSMLVE ENTDKGGRLK AEHIRNPLQT QLNWQIPVQD 300 NLPLPKGHVD LFSLSGMADD TDLALFDQNA QDNFETFSSL LGSENQQPFG ISYQAPPSSM 360 ESEFIPVKKS LLRSEESLKK VDSFSRWASK ELGEMEDLQM QSSRGDIAWT TVECETAAAG 420 ISLSPSLSED QRFTIVDFWP KCAQTDAEVE VMVIGTFLLS PQEVTKYNWS CMFGEVEVPA 480 EILVDGVLCC HAPPHTAGQV PFYVTCSNRF ACSEVREFDF LSGSTQKIDA TDVYGTYTNE 540 ASLQLRFEKM LAHRNFVHEH HIFEDVGEKR RKISKIMSLK EEKEYLLSGT YQRDSIKQEP 600 KGQLFREQFE EELYIWLIHK VTEEGKGPNI LDEDGQGILH FVAALGYDWA IRPMLAAGVN 660 INFRDANGWS ALHWAAFSGR EETVAVLVSL GADAGALTDP SPELPLGKTA ADLAYANGHR 720 GISGFLAESS LTSYLEKLTV DSKENSPANS SGAKAVQTVS ERTAAPMSYG DVPEKLSLKD 780 SLTAVRNATQ AADRLHQVFR MQSFQRKQLS DIGDDDKIDI SDKLAVSFAA SKTKNPGQGD 840 VSLSSAATHI QKKYRGWKKR KEFLLIRQRI VKIQAHVRGH QVRKQYRTVI WSVGLLEKII 900 LRWRRKGNGL RGFKRNAVAK TVEPEPPVSA ICPTIPQEDE YDYLKEGRKQ TEERLEKALT 960 RVKSMVQYPE ARDQYRRLLT VVEGFRENEA SSSASINNKE EEEVNCEEDD FIDIDSLLND 1020 DTLMMSISP* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro (PubMed:11925432). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance (PubMed:19270186). Involved in freezing tolerance in association with CAMTA2 and CAMTA3. Contributes together with CAMTA2 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved in drought stress responses by regulating several drought-responsive genes (PubMed:23547968). Involved in auxin signaling and responses to abiotic stresses (PubMed:20383645). Activates the expression of the V-PPase proton pump AVP1 in pollen (PubMed:14581622). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:11925432, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:23547968, ECO:0000269|PubMed:23581962, ECO:0000305|PubMed:11925432}. | |||||
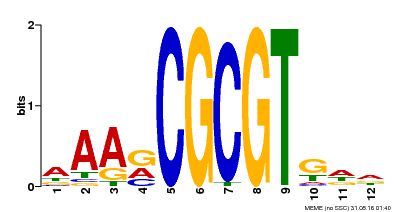
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00501 | DAP | Transfer from AT5G09410 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.13692s0022.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by UVB, wounding, ethylene and methyl jasmonate (PubMed:12218065). Induced by salt stress and heat shock (PubMed:12218065, PubMed:20383645). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK228740 | 0.0 | AK228740.1 Arabidopsis thaliana mRNA for Calmodulin-binding transcription activator 1, complete cds, clone: RAFL16-07-H14. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020878491.1 | 0.0 | calmodulin-binding transcription activator 1 isoform X4 | ||||
| Swissprot | Q9FY74 | 0.0 | CMTA1_ARATH; Calmodulin-binding transcription activator 1 | ||||
| TrEMBL | D7M234 | 0.0 | D7M234_ARALL; Calmodulin-binding transcription activator 1 | ||||
| STRING | Bostr.13129s0055.1.p | 0.0 | (Boechera stricta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4082 | 24 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G09410.2 | 0.0 | ethylene induced calmodulin binding protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.13692s0022.1.p |



