 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.16782s0002.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 249aa MW: 27665 Da PI: 8.4762 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 84.5 | 1.1e-26 | 1 | 46 | 10 | 55 |
G2-like 10 LHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
LH+rFv+av++LGG++kAtPk++l+lm++kgLtl+h+kSHLQkYRl
Araha.16782s0002.1.p 1 LHDRFVDAVAKLGGADKATPKSVLKLMGLKGLTLYHLKSHLQKYRL 46
9********************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| TIGRFAMs | TIGR01557 | 1.5E-18 | 1 | 47 | IPR006447 | Myb domain, plants |
| SuperFamily | SSF46689 | 9.32E-11 | 1 | 49 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-23 | 1 | 48 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 11.463 | 1 | 49 | IPR017930 | Myb domain |
| Pfam | PF00249 | 1.1E-5 | 2 | 45 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 4.6E-19 | 97 | 141 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 249 aa Download sequence Send to blast |
LHDRFVDAVA KLGGADKATP KSVLKLMGLK GLTLYHLKSH LQKYRLGQQQ GKKQNRTEQN 60 KENAGSSYVH FDNCSQGGIS NESRFDSHQR QSGNVPFAEA MRHQVDAQQR FQEQLEVQKK 120 LQMRMEAQGK YLLTLLEKAQ QSIPCGNAGE TDKGQFSDFN LALSRLVGSD HKNQKAGLVA 180 NISHLNADSS EDFRLCGEQE KIETGDACVK PESGFVHFDL NSKSGYDLLN CGKYGIEVKP 240 NMIADRHQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 6e-15 | 1 | 45 | 10 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 6e-15 | 1 | 45 | 10 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 6e-15 | 1 | 45 | 10 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 6e-15 | 1 | 45 | 10 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00544 | DAP | Transfer from AT5G45580 | Download |
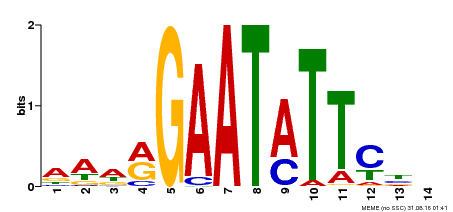
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.16782s0002.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT012003 | 0.0 | BT012003.1 Arabidopsis thaliana At5g45580 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002863468.1 | 1e-177 | myb family transcription factor PHL11 | ||||
| Swissprot | C0SVS4 | 1e-172 | PHLB_ARATH; Myb family transcription factor PHL11 | ||||
| TrEMBL | D7MTE0 | 1e-175 | D7MTE0_ARALL; Uncharacterized protein | ||||
| STRING | fgenesh1_pg.C_scaffold_8000236 | 1e-176 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM12495 | 25 | 30 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45580.1 | 1e-175 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.16782s0002.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



