 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.17006s0006.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1011aa MW: 113257 Da PI: 6.6516 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 12.9 | 0.00034 | 893 | 918 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
y+C C++sFs+ +L+ H r+ +
Araha.17006s0006.1.p 893 YQCDmeGCTMSFSSEKQLTLHKRNiC 918
99********************9877 PP
| |||||||
| 2 | zf-C2H2 | 15.5 | 4.8e-05 | 918 | 940 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk F ++ +L++H+r+H
Araha.17006s0006.1.p 918 CPvkGCGKNFFSHKYLVQHQRVH 940
9999*****************99 PP
| |||||||
| 3 | zf-C2H2 | 12.4 | 0.00047 | 976 | 1002 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C Cg++F+ s++ rH r+ H
Araha.17006s0006.1.p 976 YVCAepGCGQTFRFVSDFSRHKRKtgH 1002
899999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51184 | 16.888 | 1 | 91 | IPR003347 | JmjC domain |
| Pfam | PF02373 | 5.6E-17 | 19 | 74 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 1.87E-11 | 22 | 110 | No hit | No description |
| SMART | SM00355 | 7.5 | 893 | 915 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 13.173 | 916 | 945 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.01 | 916 | 940 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 8.3E-7 | 917 | 944 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 918 | 940 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.26E-10 | 933 | 974 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 6.9E-9 | 945 | 969 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0015 | 946 | 970 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.928 | 946 | 975 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 948 | 970 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.27E-8 | 964 | 998 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.3E-9 | 970 | 999 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.62 | 976 | 1002 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.032 | 976 | 1007 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 978 | 1002 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1011 aa Download sequence Send to blast |
EVVRVHGYGG ELNPLVTFST LGEKTTVMSP EVFVKAGIPC CRLVQNPGDF VVTFPGAYHS 60 GFSHGFNFGE ASNIATPEWL RMAKDAAIRR AAINYPPMVS HLQLLYDFAL ALGSRVPTSI 120 NAKPRSSRLK DKLRSEGERL TKKLFVQNII HNNELLYSLG KGSPVALLPQ SSSDISVCSD 180 LRIGSHLITN QENPIQLKSE DLSSDSVMVG LSNGLKDTVS VKEKFTSLCE RSRNHLASRE 240 KDTQEILSDA ERRKNDGAVA LSDQRLFSCV TCGVLSFDCV AIVQPKEAVA RYLMSADCSF 300 FNDWTAASGS ANLGQAARSL HPLSTEKHDV NYFYNVPVQT MDHSMKTGDQ RTSTTSLTMA 360 HKDNGALGLL ASAYGDSSDS EEEGQKGLDV PTSEGETKKY DKEDPDGNEE ARDGRTSDFN 420 SQRLTSEQNR LSKGGNSSLL EIALPFIPRS DDDSCRLHVF CLEHAAEVEQ QLRPFGGIHI 480 MLLCHPEYPR IEAEAKIVAE ELVINHEWND TEFRNVTRED EETIQAALDN VEAKGGNSDW 540 TVKLGINLSY SAILSRSPLY SKQMPYNSVI YNAFGRSSPS TSSPLKPEVS GKRSSRQRKY 600 VVGKWCGKVW MSHQREDGVS DDTSEDHSYK QQWRASGNEE ESYFETGNTV SGDSSNQMSD 660 QQLKGIRRHR GVKEFESDDE VSDRSLGEEY TVRGCAASES SMENGFQQSM YDDDDDDDDD 720 DINRHPRGVP RSERITVFRN PVSYDSEENG IYHQRGRVSR SNRQANRMGG EYDSEENSLE 780 EQDFCSTGKR QTRSTAKRKV KIETVRSPRD TKGRTLQEFA SGKKNEELDS YMEGPSTRLR 840 VRNLKPLRAA SETKPKKIGK KRSGNASFSR VATEEDVEED EEAENEEEEC AAYQCDMEGC 900 TMSFSSEKQL TLHKRNICPV KGCGKNFFSH KYLVQHQRVH SDDRPLKCPW KGCKMTFKWA 960 WSRTEHIRVH TGARPYVCAE PGCGQTFRFV SDFSRHKRKT GHSVKKTKKR * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6a57_A | 2e-80 | 890 | 1010 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6a58_A | 2e-80 | 890 | 1010 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6a59_A | 2e-80 | 890 | 1010 | 20 | 140 | Lysine-specific demethylase REF6 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
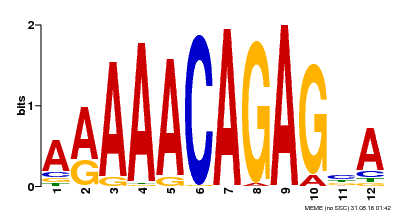
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.17006s0006.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY664499 | 0.0 | AY664499.1 Arabidopsis thaliana relative of early flowering 6 (REF6) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002875902.1 | 0.0 | lysine-specific demethylase REF6 isoform X2 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | D7LRW5 | 0.0 | D7LRW5_ARALL; Uncharacterized protein | ||||
| STRING | fgenesh2_kg.5__1196__AT3G48430.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5259 | 28 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.17006s0006.1.p |



