 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.17613s0007.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1056aa MW: 117586 Da PI: 6.3868 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 188.3 | 7.8e-59 | 19 | 136 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahse 89
l+e ++rwl++ ei++iL n++k+++++e+++rp+sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg+++vl+cyYah+e
Araha.17613s0007.2.p 19 LSEaQHRWLRPAEICEILRNHQKFHIASEPPNRPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSIDVLHCYYAHGE 107
45559************************************************************************************ PP
CG-1 90 enptfqrrcywlLeeelekivlvhylevk 118
+n++fqrrcyw+Le++l +iv+vhylevk
Araha.17613s0007.2.p 108 DNENFQRRCYWMLEQDLMHIVFVHYLEVK 136
**************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 86.275 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.6E-87 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 8.4E-52 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 6.72E-13 | 468 | 552 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 1.12E-19 | 642 | 760 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 1.4E-8 | 645 | 726 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 20.022 | 649 | 760 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.1E-18 | 650 | 761 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.23E-15 | 655 | 758 | No hit | No description |
| SMART | SM00248 | 1000 | 666 | 695 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.458 | 666 | 698 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.0029 | 699 | 728 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 10.873 | 699 | 731 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 0.32 | 874 | 896 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 9.56E-8 | 875 | 924 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| Pfam | PF00612 | 0.002 | 876 | 895 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.858 | 876 | 904 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0092 | 897 | 919 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.304 | 898 | 922 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.6E-4 | 900 | 919 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1056 aa Download sequence Send to blast |
MADRGSFGFA PRLDIKQLLS EAQHRWLRPA EICEILRNHQ KFHIASEPPN RPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHEK LKVGSIDVLH CYYAHGEDNE NFQRRCYWML 120 EQDLMHIVFV HYLEVKGNRM STGGTKENHS NSLSGTGSVN FDSTATRSSI LSPLCEDADS 180 GDSRQASSSL QQNPEPQTVV PQIMHHQNAS TMNSYNTTSV LGNRDGWTSA PGIGIVSQVH 240 GNRVKESNSQ RSGDVPAWDA SFENSLARCQ NLPYNAPLTQ TQPSTIGLIP MEGKTEKGSL 300 LTSEHLRNPL QNQIPVQESV PLQKWPMDSH SGMTDATDLA LFGQGAHENF GTFSSLLGSQ 360 NQQPSIFQAP FTNNEAAYIP KLGPEDLIYE ASANQTLPLR KALLKKEDSL KKVDSFSRWV 420 SQELGEMEDL QMQSSSGGIA WTSVECENAA AGSSLSPSLS EDQRFTMIDF WPKWTQTDSE 480 VEVMVIGTFL LSPQEVTSYS WSCMFGEVEV PADILVDGVL CCHAPPHEVG RVPFYITCSD 540 RFSCSEVREF DFLPGSTTKL NATDIYGANT IETSLHVRFE NLLALRSSVQ EHHVFENVGE 600 KRRKISKIML LKDEKESLLP GTVEKDLTEL EAKERLIREE FENKLCLWLI HKVTEEGKGP 660 NILDEDGQGV LHLAAALGYD WAIKPILAAG VSINFRDANG WSALHWAAFS GREDTVAVLV 720 SLGADAGALA DPSPEHPLGK TAADLAYGNG HRGISGFLAE SSLTSYLEKL TVDAKENSSA 780 DSSGAKAVLT VAERTATPMS YGDVPETLSM KDSLTAVFNA TQAADRLHQV FRMQSFQRKQ 840 LSELGGDNKF DISDELAVSF AAAKTKKPGH SNGAVHAAAV QIQKKYRGWK KRKEFLLIRQ 900 RIVKIQAHVR GHQVRKQYRA IIWSVGLLEK IILRWRRKGS GLRGFKRDTI TKPTEPVCPA 960 PQEDDYDFLK EGRKQTEERL KKALTRVKSM AQYPEARAQY RRLLTVVEGF RENEASSSSA 1020 LKNNTEEAAN YNEEDDLIDI DSLLDDDTFM SLAFE* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
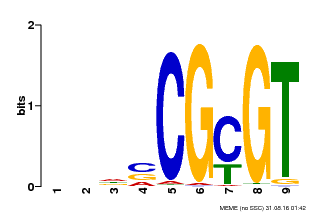
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.17613s0007.2.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK229403 | 0.0 | AK229403.1 Arabidopsis thaliana mRNA for Calmodulin-binding transcription activator 2, complete cds, clone: RAFL16-66-C08. | |||
| GenBank | BT010874 | 0.0 | BT010874.1 Arabidopsis thaliana At5g64220 gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020883660.1 | 0.0 | calmodulin-binding transcription activator 2 isoform X1 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | D7MR88 | 0.0 | D7MR88_ARALL; Calmodulin-binding protein | ||||
| STRING | scaffold_803130.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.17613s0007.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



