 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.17880s0001.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 674aa MW: 78419.3 Da PI: 6.9854 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 90.9 | 1.7e-28 | 66 | 154 | 2 | 91 |
FAR1 2 fYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkktekerrtraetrtgCkaklkvkkekdgkwevtkleleHnHela 90
fY+eYAk++GF++++++s++sk+++ +++++f Cs++g + e++++ +++r++++++t+Cka+++vk++ dgkw +++++++HnHel
Araha.17880s0001.1.p 66 FYQEYAKSMGFTTSIKNSRRSKKTKDFIDAKFACSRYGVTPESESS-GSSSRRSTVKKTDCKASMHVKRKPDGKWIIHEFVKDHNHELL 153
9***************************************999998.9999999*********************************97 PP
FAR1 91 p 91
p
Araha.17880s0001.1.p 154 P 154
5 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 2.8E-26 | 66 | 154 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 1.0E-25 | 275 | 368 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 8.939 | 555 | 591 | IPR007527 | Zinc finger, SWIM-type |
| Pfam | PF04434 | 7.5E-7 | 557 | 590 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 3.1E-7 | 566 | 593 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010018 | Biological Process | far-red light signaling pathway | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 674 aa Download sequence Send to blast |
MDLQENLVSD AGDEHMVDIV VEPHSNRDIG IVDEFNLGGD VGFSGDLDLE PRNGIDFETH 60 EAAYIFYQEY AKSMGFTTSI KNSRRSKKTK DFIDAKFACS RYGVTPESES SGSSSRRSTV 120 KKTDCKASMH VKRKPDGKWI IHEFVKDHNH ELLPALAYHF RIQRNVKLAE KNNIDILHAV 180 SERTKKMYVE MSRQSGGYKN IGPLLQTDVS SQVDKGRYLA LEDGDSQVLL EYFKRIKKEN 240 PKFFYAIDLN EDQRLRNLFW ADAKSRDDYL SFNDVVSFDT TYVKFNDKLP LALFIGVNHH 300 SQPMLLGCAL VADESMETFV WLIKTWLRAM SGRAPKVILT DQDKFLMSAV SELLPNTRHC 360 FALWHVLEKI PEYFSHVMKQ HENFLQKFNK CIFRSWTDDE FDMRWWKMVS QFGLENDEWL 420 LWLHEHRQKW VPTFMSNVFL AGMSTSQRSE SVNSFFDKYV HKKITLKEFL RQYGAILQNR 480 YEEESVADFD TCHKQPALKS PSPWEKQMAT TYTHTIFKKF QVEVLGVVAC HPRKEKEDEN 540 MATFRVQDCE KDDDFLVTWS KTKSELCCFC RMFEYKGFLC RHALMILQMC GFASIPPRYI 600 LKRWTKDAKS GVLAGEGADQ IQTRVQRYND LCSRAIELSE EGCVSEENYN IVLRTLVETL 660 KNCVDMNNAR NNIT |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00434 | DAP | Transfer from AT4G15090 | Download |
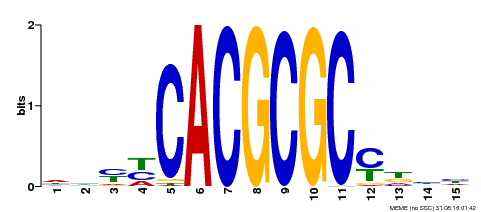
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.17880s0001.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. {ECO:0000269|PubMed:18033885}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF159587 | 0.0 | AF159587.1 Arabidopsis thaliana far-red impaired response protein (FAR1) mRNA, complete cds. | |||
| GenBank | CP002687 | 0.0 | CP002687.1 Arabidopsis thaliana chromosome 4 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020874483.1 | 0.0 | protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| Refseq | XP_020874484.1 | 0.0 | protein FAR-RED IMPAIRED RESPONSE 1 isoform X2 | ||||
| Swissprot | Q9SWG3 | 0.0 | FAR1_ARATH; Protein FAR-RED IMPAIRED RESPONSE 1 | ||||
| TrEMBL | A0A178UY59 | 0.0 | A0A178UY59_ARATH; FAR1 | ||||
| STRING | AT4G15090.1 | 0.0 | (Arabidopsis thaliana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM118 | 26 | 253 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G15090.1 | 0.0 | FAR1 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.17880s0001.1.p |



