 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.21147s0001.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 541aa MW: 59930.6 Da PI: 7.3221 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 55.5 | 9.9e-18 | 284 | 333 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h++ E+rRR++iN++f+ Lr+l+P++ ++K +Ka+ L +++eYI+ Lq
Araha.21147s0001.1.p 284 RSKHSATEQRRRSKINDRFQMLRQLIPNS----DQKRDKASFLLEVIEYIQFLQ 333
889*************************9....9*******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 1.0E-23 | 281 | 353 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.225 | 282 | 332 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 6.8E-19 | 283 | 352 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.3E-15 | 284 | 333 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 7.56E-15 | 284 | 335 | No hit | No description |
| SMART | SM00353 | 3.4E-13 | 288 | 338 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 541 aa Download sequence Send to blast |
MELPQPRPFK TEGRKPTHDF LSLCSHSTVH PDPKPTPPSS QGSHLKTHDF LQPLECVAAK 60 EEVSRINTTT TASEKPPPPA PPPPLQHVLP GGIGTYTISP IPYFHHHHQR IPKPELSPPM 120 MFNAAAQASG GNERNIVDEN SNSNCSSYAA AASGFTLWDE SASGKKGQTR KENNVVERVN 180 MRADVAATVG QWPVVERRSQ SLTNNHMSGF SSRSSSQGSG LKSQSFMDMI RSAKGSSQDD 240 DLDDEEDFIM KKESSSTSQS HRVDLRVKAD VRGSANDQKL NTPRSKHSAT EQRRRSKIND 300 RFQMLRQLIP NSDQKRDKAS FLLEVIEYIQ FLQEKADKYE TSYQGWNHEP AKLLNWQSNN 360 QQLVPEGVAF APKMEEEKNN IPVSVLATAQ GVVIDHPTTA TTSPFPLSIQ SNSFFSPVIA 420 GNPVPQFHTR VTSSETVEPS PSSRNQTQPL KEEEEVEDEE VVEGNIRISS VYSQGLVKTL 480 REALENSGVD LTKASISVEI ELAKQSSSSF KGHEVREPVS RTRNDDVKQT RKPKRLKTGQ 540 * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. Transcription factor that bind specifically to the DNA sequence 5'-CANNTG-3'(E box). Can bind individually to the promoter as a homodimer or synergistically as a heterodimer with BZR2/BES1. Does not itself activate transcription but enhances BZR2/BES1-mediated target gene activation. {ECO:0000269|PubMed:15680330}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
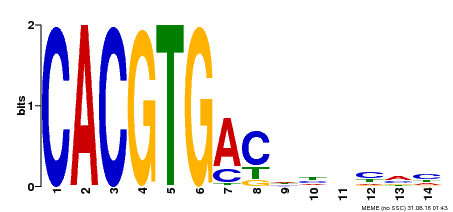
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.21147s0001.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by cold treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353313 | 0.0 | AK353313.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-34-P07. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002873342.1 | 0.0 | transcription factor BIM1 isoform X1 | ||||
| Swissprot | Q9LEZ3 | 0.0 | BIM1_ARATH; Transcription factor BIM1 | ||||
| TrEMBL | D7M188 | 0.0 | D7M188_ARALL; Uncharacterized protein | ||||
| STRING | scaffold_600819.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6967 | 27 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.4 | 0.0 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.21147s0001.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



