 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.2135s0007.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 435aa MW: 49003.5 Da PI: 8.5522 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56.7 | 5.3e-18 | 93 | 139 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEd +l+++v+q+G + W++I++++ gR +kqc++rw+++l
Araha.2135s0007.1.p 93 KGQWTAEEDRKLIRLVRQHGERKWALISEKLE-GRAGKQCRERWHNHL 139
799*****************************.*************97 PP
| |||||||
| 2 | Myb_DNA-binding | 46.2 | 1e-14 | 166 | 207 | 2 | 45 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ W++eE+ lv+ + + G++ W+ Ia+ ++ gRt++++k++w+
Araha.2135s0007.1.p 166 DGWSEEEERVLVESHMRIGNK-WAEIAKLIP-GRTENSIKNHWN 207
56*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 29.445 | 88 | 143 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.67E-28 | 90 | 206 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.3E-16 | 92 | 141 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.0E-16 | 93 | 139 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-23 | 94 | 148 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 9.55E-16 | 96 | 139 | No hit | No description |
| PROSITE profile | PS51294 | 17.748 | 160 | 214 | IPR017930 | Myb domain |
| SMART | SM00717 | 5.8E-13 | 164 | 212 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.1E-13 | 167 | 207 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.18E-10 | 168 | 210 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-16 | 168 | 208 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 435 aa Download sequence Send to blast |
MRYVFGTFLY GQKNDVLCSK KQECSRDRPI VKTKMSIETR SDNKENTTFG PTREKHVVLN 60 GGNRSPIGEV VVRSAARDYQ NSTKKRPYKN LIKGQWTAEE DRKLIRLVRQ HGERKWALIS 120 EKLEGRAGKQ CRERWHNHLR PDIKGSDICP SISTFLPKQT KNNQKDGWSE EEERVLVESH 180 MRIGNKWAEI AKLIPGRTEN SIKNHWNATK RRQNSKRKHK RESNADNNDT DVSPSAKRPC 240 ILQDYIKSIG SNDINKNNDE NTISVLSTPN LDQIYSDGDS ASSMLGDPYD EELVYLQNIF 300 ANHPISLENI GLSQTSDEIT QSSASGFMTK KPNPNLHNNV GTHHQEAAIT APVNTPHLAS 360 DIYLSYLLNG TTSSYSDTHF PSSSSSTSST TVEHGGHNEF LEPQANSTSE RREMDLIEML 420 SGSIQGSNIC FPLF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1mse_C | 8e-40 | 91 | 213 | 2 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 8e-40 | 91 | 213 | 2 | 104 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 209 | 220 | KRRQNSKRKHKR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor required for female gametophyte fertility. Acts redundantly with MYB64 to initiate the FG5 transition during female gametophyte development. The FG5 transition represents the switch between free nuclear divisions and cellularization-differentiation in female gametophyte, and occurs during developmental stage FG5. {ECO:0000269|PubMed:24068955}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00567 | DAP | Transfer from AT5G58850 | Download |
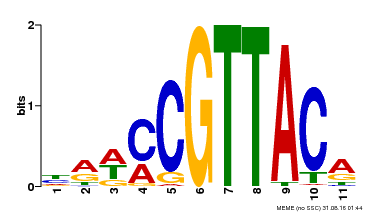
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.2135s0007.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB016885 | 0.0 | AB016885.1 Arabidopsis thaliana genomic DNA, chromosome 5, TAC clone:K19M22. | |||
| GenBank | CP002688 | 0.0 | CP002688.1 Arabidopsis thaliana chromosome 5 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020887723.1 | 0.0 | myb-like protein Q | ||||
| Swissprot | Q9FIM4 | 0.0 | MY119_ARATH; Transcription factor MYB119 | ||||
| TrEMBL | D7L578 | 0.0 | D7L578_ARALL; MYB119 | ||||
| STRING | fgenesh2_kg.3__2684__AT5G58850.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4265 | 26 | 56 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G58850.1 | 0.0 | myb domain protein 119 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.2135s0007.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



