 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.23701s0002.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 326aa MW: 37261.7 Da PI: 5.7461 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 110.1 | 1.6e-34 | 18 | 110 | 2 | 103 |
HHHHHHHHHCTGGGTTTSEESSSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTTXTTSEEEEESXXXXXX CS
HSF_DNA-bind 2 FlkklyeiledeelkeliswsengnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekkskskekiweFkhksFkkgk 90
F+ k+y++++d+ ++ li+w nsf+v+d+ +f++++Lp yFkh+nf+SFvRQLn+YgF+kv+ ++ weF++++F +g+
Araha.23701s0002.1.p 18 FIVKTYQMVNDPLTDWLITWGPAHNSFIVVDPLDFSQRILPAYFKHNNFSSFVRQLNTYGFRKVDPDR---------WEFANEHFLRGQ 97
9********************999*****************************************999.........************ PP
XXXXXXXXXXXXX CS
HSF_DNA-bind 91 kellekikrkkse 103
k+ll++i r+k++
Araha.23701s0002.1.p 98 KHLLKNIARRKHA 110
**********975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00415 | 1.9E-53 | 14 | 107 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| SuperFamily | SSF46785 | 1.5E-32 | 15 | 107 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 9.7E-37 | 15 | 107 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| PRINTS | PR00056 | 9.3E-18 | 18 | 41 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Pfam | PF00447 | 1.2E-29 | 18 | 107 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 9.3E-18 | 56 | 68 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PROSITE pattern | PS00434 | 0 | 57 | 81 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 9.3E-18 | 69 | 81 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006970 | Biological Process | response to osmotic stress | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 326 aa Download sequence Send to blast |
MEDDNGNNTN NNNVIAPFIV KTYQMVNDPL TDWLITWGPA HNSFIVVDPL DFSQRILPAY 60 FKHNNFSSFV RQLNTYGFRK VDPDRWEFAN EHFLRGQKHL LKNIARRKHA RGMYGQDLED 120 GEIVREIERL KEEQRELEAE IQRMNQRIEA TEKRPEQMMA FLYKVVEDPD LLPRMMLEKE 180 RTKQQVSDKK KRRVTMSTVK AEEEEVEEDE ARVFRVISSS TPSPSSTENL YRNHSPDGWV 240 VPMTQGQFGN YETGLVANSM LSNSTSSTSS SLTSTFSLPE SVNGGGCGNI QGERRYKETA 300 TFGGVVESNP PTTPPYPFSL FRGGF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5d5u_B | 2e-25 | 15 | 107 | 26 | 129 | Heat shock factor protein 1 |
| 5d5v_B | 2e-25 | 15 | 107 | 26 | 129 | Heat shock factor protein 1 |
| 5d5v_D | 2e-25 | 15 | 107 | 26 | 129 | Heat shock factor protein 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 188 | 192 | KKKRR |
| 2 | 188 | 193 | KKKRRV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
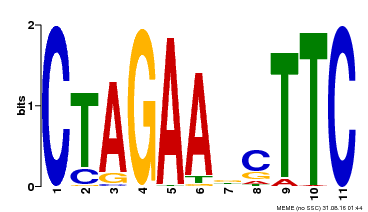
| UniProt | Transcriptional regulator that specifically binds DNA sequence 5'-AGAAnnTTCT-3' known as heat shock promoter elements (HSE). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00379 | DAP | Transfer from AT3G24520 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.23701s0002.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HQ322382 | 0.0 | HQ322382.1 Arabidopsis thaliana heat shock factor (HSF) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020889030.1 | 0.0 | heat stress transcription factor C-1 | ||||
| Swissprot | Q9LV52 | 0.0 | HSFC1_ARATH; Heat stress transcription factor C-1 | ||||
| TrEMBL | D7L548 | 0.0 | D7L548_ARALL; Uncharacterized protein (Fragment) | ||||
| STRING | fgenesh2_kg.3__2665__AT3G24520.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6775 | 27 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G24520.1 | 0.0 | heat shock transcription factor C1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.23701s0002.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



