 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.2597s0023.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 584aa MW: 65299.4 Da PI: 6.8231 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 43.5 | 7.9e-14 | 271 | 330 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pse.ng..kr.krfslgkfgtaeeAakaaiaarkkleg 55
s y+GV++++++gr++A+++d + g ++ ++++lg ++ +++Aa+a++ a++k+++
Araha.2597s0023.1.p 271 SIYRGVTRHRWTGRYEAHLWDnSCRrEGqaRKgRQVYLGGYDKEDKAARAYDLAALKYWN 330
57*******************666664478446**********99*************97 PP
| |||||||
| 2 | AP2 | 49.6 | 9.7e-16 | 373 | 424 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s y+GV++++++grW A+I + +k +lg+f t+eeAa+a++ a+ k++g
Araha.2597s0023.1.p 373 SIYRGVTRHHQQGRWQARIGRVAG---NKDLYLGTFATEEEAAEAYDIAAIKFRG 424
57****************988532...5************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 3.79E-16 | 271 | 339 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 8.7E-11 | 271 | 330 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.07E-20 | 271 | 340 | No hit | No description |
| PROSITE profile | PS51032 | 18.545 | 272 | 338 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.1E-25 | 272 | 344 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.6E-14 | 272 | 338 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.3E-6 | 273 | 284 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 4.1E-10 | 373 | 424 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.31E-11 | 373 | 432 | No hit | No description |
| SuperFamily | SSF54171 | 1.11E-17 | 373 | 433 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 19.111 | 374 | 432 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 7.9E-31 | 374 | 438 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 5.5E-18 | 374 | 432 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.3E-6 | 414 | 434 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010080 | Biological Process | regulation of floral meristem growth | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0035265 | Biological Process | organ growth | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0060772 | Biological Process | leaf phyllotactic patterning | ||||
| GO:0060774 | Biological Process | auxin mediated signaling pathway involved in phyllotactic patterning | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 584 aa Download sequence Send to blast |
MAPMTNWLTF SLSPMEMLRS SDQSQFVSYD ASSAASSSPY LLDNFYGWTN QKPQEFFKEE 60 AQLAAASMAD STILTTFVDP QSHSQNHIPK LEDFLGDSSS IVRYSDNSQT DTQDSSLTQI 120 YDPRHHHHHN HNQTGFYSDH HDFKTMAGFQ TAFSTNSGSE VDDSASIGRT HLTGEYLGHV 180 VESSGPELGF HGGSTGALSL GVNVNNTNHR NDNDSNQITN HHYRGNNNNG DRINNNENEK 240 TDSEKEKAVV AVETSDCSNK KIADTFGQRT SIYRGVTRHR WTGRYEAHLW DNSCRREGQA 300 RKGRQVYLGG YDKEDKAARA YDLAALKYWN ATATTNFPIT NYSKEVEEMK HMTKQEFIAS 360 LRRKSSGFSR GASIYRGVTR HHQQGRWQAR IGRVAGNKDL YLGTFATEEE AAEAYDIAAI 420 KFRGINAVTN FEMNRYDVEA IMKSALPIGG AAKRLKLSLE AASSEQKPIL GHQLHHFQQQ 480 QQQQQQLQLQ SSPNHSSINF ALCPNSAAQS QQIIPCGIPF EAAALYHHQQ QQQQQQQQNF 540 FQHFPANAAS DSTGSNNNSN VQGTMGLMAP NPAEFFLWPN QSY* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00504 | DAP | Transfer from AT5G10510 | Download |
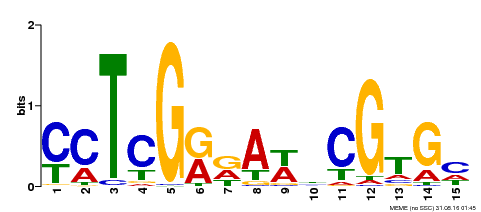
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.2597s0023.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY585683 | 0.0 | AY585683.1 Arabidopsis thaliana clone at5g10510 AP2/EREBP transcription factor mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020876725.1 | 0.0 | AP2-like ethylene-responsive transcription factor AIL6 isoform X1 | ||||
| Refseq | XP_020876726.1 | 0.0 | AP2-like ethylene-responsive transcription factor AIL6 isoform X1 | ||||
| Swissprot | Q52QU2 | 0.0 | AIL6_ARATH; AP2-like ethylene-responsive transcription factor AIL6 | ||||
| TrEMBL | D7M2X9 | 0.0 | D7M2X9_ARALL; Uncharacterized protein | ||||
| STRING | scaffold_601064.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3333 | 27 | 65 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G10510.2 | 0.0 | AINTEGUMENTA-like 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.2597s0023.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



