 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.30828s0011.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 197aa MW: 22072.3 Da PI: 5.4844 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 68 | 9e-22 | 9 | 56 | 1 | 48 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEE CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklye 48
krienks rqvtfskRrng++ KA LS+LC+ vav+++s +gkly+
Araha.30828s0011.1.p 9 KRIENKSSRQVTFSKRRNGLIDKARQLSILCESSVAVVVVSASGKLYD 56
79********************************************97 PP
| |||||||
| 2 | K-box | 35.4 | 4.4e-13 | 95 | 164 | 29 | 98 |
K-box 29 enLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkkl 98
e L++ q +l ++++ s+ L +Le+qLe++l+ R++K el++e ++ l++ke l+een+ L +++
Araha.30828s0011.1.p 95 ELLETVQSKLEEPSVDNVSVDSLISLEEQLETALSVSRARKAELMMEYVKSLKEKEMLLREENQVLASQM 164
66777777788889999************************************************99876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF55455 | 1.44E-25 | 1 | 73 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 28.202 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 4.7E-33 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.0E-23 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 2.1E-21 | 10 | 55 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.0E-23 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.0E-23 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS51297 | 11.333 | 80 | 170 | IPR002487 | Transcription factor, K-box |
| Pfam | PF01486 | 1.5E-10 | 95 | 164 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 197 aa Download sequence Send to blast |
MGRRKIEIKR IENKSSRQVT FSKRRNGLID KARQLSILCE SSVAVVVVSA SGKLYDSSSG 60 DDISKIIDRY EIQHADELKA LDLAEKIQNY LPHKELLETV QSKLEEPSVD NVSVDSLISL 120 EEQLETALSV SRARKAELMM EYVKSLKEKE MLLREENQVL ASQMGKNTLL AIEDERGMLP 180 ESSSGNKIPE TLPLLK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6bz1_A | 3e-15 | 1 | 70 | 1 | 69 | MEF2 CHIMERA |
| 6bz1_B | 3e-15 | 1 | 70 | 1 | 69 | MEF2 CHIMERA |
| 6bz1_C | 3e-15 | 1 | 70 | 1 | 69 | MEF2 CHIMERA |
| 6bz1_D | 3e-15 | 1 | 70 | 1 | 69 | MEF2 CHIMERA |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the negative regulation of flowering time in both long and short days, probably through the photoperiodic and vernalization pathways. Prevents premature flowering. {ECO:0000269|PubMed:11351076, ECO:0000269|PubMed:14558654, ECO:0000269|PubMed:15695584, ECO:0000269|PubMed:18799658}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00105 | ChIP-seq | Transfer from AT1G77080 | Download |
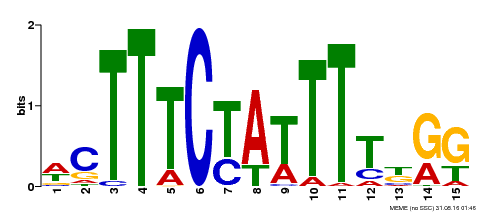
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.30828s0011.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly repressed by vernalization. Negatively regulated at the chromatin level by VIL1 through the photoperiod and vernalization pathways. Requires EARLY FLOWERING 7 (ELF7) and ELF8 to be expressed. Up-regulated by HUA2. {ECO:0000269|PubMed:11351076, ECO:0000269|PubMed:15520273, ECO:0000269|PubMed:15659097, ECO:0000269|PubMed:17114575}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF342808 | 0.0 | AF342808.1 Arabidopsis thaliana MADS affecting flowering 1 (MAF1) mRNA, complete cds. | |||
| GenBank | AK117577 | 0.0 | AK117577.1 Arabidopsis thaliana mRNA for (NM_148660) MADS affecting flowering 1(MAF1); protein id: At1g77080.4, supported by cDNA: 92459., supported by cDNA: gi_13649968, supported by cDNA: gi_16580104 [Arabidopsis thaliana] gb|AAK37527.1|AF342808_1 (AF342808) MADS affecting flowering 1 [Arabidopsis thaliana] gb|AAK54440.1| (AY034083) MADS box FLC1-like nuclear protein [Arabidopsis thaliana] gb|AAM67028.1| (AY088709) MADS affecting flowering 1 [Arabidopsis thaliana], complete cds, clone: RAFL17-22-J12. | |||
| GenBank | AK175247 | 0.0 | AK175247.1 Arabidopsis thaliana mRNA, complete cds, clone: RAFL21-67-N06. | |||
| GenBank | AK176270 | 0.0 | AK176270.1 Arabidopsis thaliana mRNA, complete cds, clone: RAFL23-17-J21. | |||
| GenBank | AK227948 | 0.0 | AK227948.1 Arabidopsis thaliana mRNA for hypothetical protein, complete cds, clone: RAFL14-50-E05. | |||
| GenBank | AY034083 | 0.0 | AY034083.1 Arabidopsis thaliana MADS box FLC1-like nuclear protein (FK1) mRNA, complete cds. | |||
| GenBank | AY088709 | 0.0 | AY088709.1 Arabidopsis thaliana clone 92459 mRNA, complete sequence. | |||
| GenBank | BT004598 | 0.0 | BT004598.1 Arabidopsis thaliana At1g77080 gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020890340.1 | 1e-125 | agamous-like MADS-box protein AGL27 isoform X1 | ||||
| Swissprot | Q9AT76 | 1e-122 | AGL27_ARATH; Agamous-like MADS-box protein AGL27 | ||||
| TrEMBL | A0A1P8ARA3 | 1e-113 | A0A1P8ARA3_ARATH; K-box region and MADS-box transcription factor family protein | ||||
| STRING | AT1G77080.4 | 1e-120 | (Arabidopsis thaliana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM986 | 19 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G77080.4 | 1e-111 | MIKC_MADS family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.30828s0011.1.p |



