 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.3205s0010.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1032aa MW: 115801 Da PI: 5.2427 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 182.5 | 4.8e-57 | 20 | 136 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseen 91
+e ++rwl++ ei++iL+n++++++++e++t+p+sgs+++++rk++ryfrkDG++w+kkkdgktv+E+he+LK g+v+vl+cyYah+++n
Araha.3205s0010.1.p 20 SEaRHRWLRPPEICEILQNYQRFQISTEPPTTPSSGSVFMFDRKVLRYFRKDGHNWRKKKDGKTVKEAHERLKAGSVDVLHCYYAHGQDN 109
4449************************************************************************************** PP
CG-1 92 ptfqrrcywlLeeelekivlvhylevk 118
++fqrr+ywlL+eel++iv+vhylevk
Araha.3205s0010.1.p 110 ENFQRRSYWLLQEELSHIVFVHYLEVK 136
************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 82.708 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.4E-85 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 9.9E-50 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 7.5E-5 | 464 | 552 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF01833 | 5.7E-4 | 466 | 551 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 5.76E-17 | 466 | 551 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 2.8E-15 | 637 | 757 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 3.7E-15 | 642 | 757 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 4.02E-11 | 642 | 754 | No hit | No description |
| PROSITE profile | PS50297 | 16.149 | 645 | 754 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 9.164 | 695 | 727 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.51E-8 | 850 | 903 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.92 | 852 | 874 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.029 | 854 | 882 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0041 | 855 | 873 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0011 | 875 | 897 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.468 | 876 | 900 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 1.2E-4 | 878 | 897 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1032 aa Download sequence Send to blast |
MAEARRFSPD NELDVGQILS EARHRWLRPP EICEILQNYQ RFQISTEPPT TPSSGSVFMF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHER LKAGSVDVLH CYYAHGQDNE NFQRRSYWLL 120 QEELSHIVFV HYLEVKGSRV STSYNRMQRT EDAARSPQET GEALTSEHDG YASCSYNQND 180 HSNHSQTTDS ASVNGFHSPE LEDAESAYNQ HGSSIAYSHQ ELQQPATGGS LTGFDPYYQI 240 SLTPRDSYQK ELRTIPVTDS SIMVDKSKTI NSPGESNGLR NRKSIDSQTW EEILGNCGSG 300 VEALPLQPNS EHEVLDQILE SCSFTMQDFV SLQGSMVKSQ NQELNSGLTS DSTVWFQGQD 360 VELNAISNLA SNEKAPYLST MKQHLLDGAL GEEGLKKMDS FNRWMSKELG DVGVIADANE 420 SFTQSSSRTY WEEVESEDGS NGHNSRRELD GYVMSPSLSK EQLFSINDFS PSWAYVGCEV 480 VVFVTGKFLK TREETEIGEW SCMFGQTEVP ADVISNGILQ CVAPMHEAGR VPFYVTCSNR 540 LACSEVREFE YKVAESQVFD RETDDESTIN ILEARFVKLL CSKSENSSPV SGNDSHLSQL 600 SEKISLLLFE NDDQLDQMLM NEISQENMKN NLLQEFLKES LHSWLLQKIA EGGKGPSVLD 660 EGGQGVLHFA ASLGYNWALE PTIIAGVSVD FRDVNGWTAL HWAAFFGRER IIGSLIALGA 720 APGTLTDPNP DFPSGSTPSD LAYANGHKGI AGYLSEYALR AHVSLLSLND NSAETVETAP 780 SPSSSSLTDS LTAVRNATQA AARIHQVFRA QSFQKKQLKE FGDRKLGMSE ERALSMLAPK 840 THKSGRAHSD DSVQAAAIRI QNKFRGYKGR KDYLITRQRI IKIQAHVRGY QVRKNYRKII 900 WSVGILEKVI LRWRRKGAGL RGFKSEALVD KMQDGTAKEE DDDFFKQGRK QTEERLQKAL 960 ARVKSMVQYP EARDQYRRLL NVVNDIQESK VEKALENSEA TCFDDDLIDI EALLEDDDTL 1020 MLPMSSSMWT S* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3'. Binds calmodulin in a calcium-dependent manner in vitro (PubMed:12218065). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance in association with CAMTA1 and CAMTA2 (PubMed:23581962). Required for the cold-induced expression of DREB1B/CBF1, DREB1C/CBF2, ZAT12 and GOLS3 (PubMed:19270186). Involved in response to cold. Contributes together with CAMTA5 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). Involved together with CAMTA2 and CAMTA4 in the positive regulation of a general stress response (GSR) (PubMed:25039701). Involved in the regulation of GSR amplitude downstream of MEKK1 (PubMed:25157030). Involved in the regulation of a set of genes involved in defense responses against pathogens (PubMed:18298954). Involved in the regulation of both basal resistance and systemic acquired resistance (SAR) (PubMed:21900483). Acts as negative regulator of plant immunity (PubMed:19122675, PubMed:21900483, PubMed:22345509, PubMed:28407487). Binds to the promoter of the defense-related gene EDS1 and represses its expression (PubMed:19122675). Binds to the promoter of the defense-related gene NDR1 and represses its expression (PubMed:22345509). Involved in defense against insects (PubMed:23072934, PubMed:22371088). Required for tolerance to the generalist herbivore Trichoplusia ni, and contributes to the positive regulation of genes associated with glucosinolate metabolism (PubMed:23072934). Required for tolerance to Bradysia impatiens larvae. Mediates herbivore-induced wound response (PubMed:22371088). Required for wound-induced jasmonate accumulation (PubMed:23072934, PubMed:22371088). Involved in the regulation of ethylene-induced senescence by binding to the promoter of the senescence-inducer gene EIN3 and repressing its expression (PubMed:22345509). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:18298954, ECO:0000269|PubMed:19122675, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:21900483, ECO:0000269|PubMed:22345509, ECO:0000269|PubMed:22371088, ECO:0000269|PubMed:23072934, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25157030, ECO:0000269|PubMed:28351986, ECO:0000269|PubMed:28407487, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
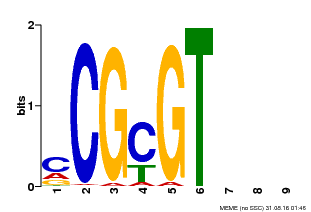
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.3205s0010.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, salt, wounding, ethylene and methyl jasmonate (PubMed:11162426, PubMed:12218065). Induced by infection with the fungal pathogen Golovinomyces cichoracearum (powdery mildew) and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 (PubMed:22345509). {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:22345509}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF506697 | 0.0 | AF506697.1 Arabidopsis thaliana calmodulin-binding transcription factor SR1 (SR1) mRNA, complete cds. | |||
| GenBank | AY510025 | 0.0 | AY510025.1 Arabidopsis thaliana ethylene-induced calmodulin-binding protein 1 mRNA, complete cds. | |||
| GenBank | BT002459 | 0.0 | BT002459.1 Arabidopsis thaliana Unknown protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020883251.1 | 0.0 | calmodulin-binding transcription activator 3 isoform X2 | ||||
| Refseq | XP_020883252.1 | 0.0 | calmodulin-binding transcription activator 3 isoform X2 | ||||
| Swissprot | Q8GSA7 | 0.0 | CMTA3_ARATH; Calmodulin-binding transcription activator 3 | ||||
| TrEMBL | D7LDE6 | 0.0 | D7LDE6_ARALL; Uncharacterized protein | ||||
| STRING | fgenesh2_kg.4__153__AT2G22300.2 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM8601 | 25 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.3205s0010.1.p |



