 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.3284s0027.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 564aa MW: 62987.5 Da PI: 5.3383 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 43.7 | 4.9e-14 | 393 | 439 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr+++P+ + K++Ka+ L A+ YIk+Lq
Araha.3284s0027.1.p 393 NHVEAERQRREKLNQRFYALRSVVPNiS------KMDKASLLGDAISYIKELQ 439
799***********************66......******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 1.3E-51 | 48 | 239 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| SuperFamily | SSF47459 | 3.66E-19 | 388 | 452 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.936 | 389 | 438 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.93E-15 | 392 | 443 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 1.3E-18 | 393 | 453 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.8E-11 | 393 | 439 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 6.3E-18 | 395 | 444 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 564 aa Download sequence Send to blast |
MNISDLGWDD EEKSVVSAVL GHLASDFLRA NSNSNQNLFL VMGTDDSLNK KLSSLVDWPN 60 SENFSWNYAI FWQQTMSRSG QQVLGWGDGC CREPTEEEES KVVRSYNFSN LGVEEETWQD 120 MRKRVLQKLH RLFGGSDEDN YALSLEKVTA TEIFFLASMY FFFNHGEGGP GRCYASGKHV 180 WLSDAVNSES DYCFRSFMAK SAGIRTIVMV PTDAGVLELG SVWSLPENIG LVKSVQALFM 240 RRVTQPLMVT SNANMSGGIH KLFGQDLNNS DRGAHAYPKK LEVRRNLDER FTPQSWEGPT 300 FGYTPQREDV KVQENVNVVV DDNNYKTQIE FAGSSVPASS NPSTNTQLEK SESCTEKRPV 360 SLLAGAGTVS VVDEKRPRKR GRKPANGREE PLNHVEAERQ RREKLNQRFY ALRSVVPNIS 420 KMDKASLLGD AISYIKELQE KVKIMEDERV RTDNSLSESN TRTVESTEVD IQAMNEEVVV 480 RVVSPLDSHP ASRIIQAMRN SNVSLMEAKL SLAEDTMFHT FVVKSNNGSD PLTKEKLIAA 540 VYPENSSTQQ PLPSSSSQVS GDI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 7e-30 | 387 | 458 | 2 | 73 | Transcription factor MYC2 |
| 5gnj_B | 7e-30 | 387 | 458 | 2 | 73 | Transcription factor MYC2 |
| 5gnj_E | 7e-30 | 387 | 458 | 2 | 73 | Transcription factor MYC2 |
| 5gnj_F | 7e-30 | 387 | 458 | 2 | 73 | Transcription factor MYC2 |
| 5gnj_G | 7e-30 | 387 | 458 | 2 | 73 | Transcription factor MYC2 |
| 5gnj_I | 7e-30 | 387 | 458 | 2 | 73 | Transcription factor MYC2 |
| 5gnj_M | 7e-30 | 387 | 458 | 2 | 73 | Transcription factor MYC2 |
| 5gnj_N | 7e-30 | 387 | 458 | 2 | 73 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 374 | 382 | KRPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator. Regulates positively abscisic acid (ABA) response. Confers drought tolerance and sensitivity to ABA. {ECO:0000269|PubMed:17828375}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00101 | PBM | Transfer from AT2G46510 | Download |
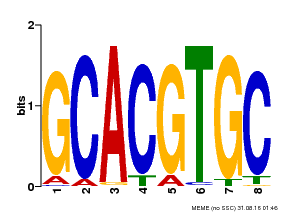
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.3284s0027.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Transiently by ABA and polyethylene glycol (PEG). {ECO:0000269|PubMed:17828375}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC006418 | 0.0 | AC006418.4 Arabidopsis thaliana chromosome 2 clone F13A10 map CIC06C03, complete sequence. | |||
| GenBank | AC006526 | 0.0 | AC006526.8 Arabidopsis thaliana chromosome 2 clone F11C10 map CIC02E07, complete sequence. | |||
| GenBank | AY094399 | 0.0 | AY094399.1 Arabidopsis thaliana At2g46510/F13A10.4 mRNA, complete cds. | |||
| GenBank | CP002685 | 0.0 | CP002685.1 Arabidopsis thaliana chromosome 2, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020884075.1 | 0.0 | transcription factor ABA-INDUCIBLE bHLH-TYPE | ||||
| Swissprot | Q9ZPY8 | 0.0 | AIB_ARATH; Transcription factor ABA-INDUCIBLE bHLH-TYPE | ||||
| TrEMBL | D7LEK3 | 0.0 | D7LEK3_ARALL; Basic helix-loop-helix family protein | ||||
| STRING | fgenesh2_kg.4__2868__AT2G46510.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3758 | 27 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46510.1 | 0.0 | ABA-inducible BHLH-type transcription factor | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.3284s0027.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



