 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.3358s0010.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | Nin-like | ||||||||
| Protein Properties | Length: 960aa MW: 105716 Da PI: 5.851 | ||||||||
| Description | Nin-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | RWP-RK | 91.9 | 4.9e-29 | 589 | 639 | 2 | 52 |
RWP-RK 2 ekeisledlskyFslpikdAAkeLgvclTvLKriCRqyGIkRWPhRkiksl 52
ek+isl++l++yF +++kdAAk+Lgvc+T++KriCRq+GI+RWP+Rkik++
Araha.3358s0010.1.p 589 EKTISLDVLQQYFTGSLKDAAKSLGVCPTTMKRICRQHGISRWPSRKIKKV 639
799**********************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF55781 | 4.71E-5 | 214 | 283 | IPR029016 | GAF domain-like |
| PROSITE profile | PS51519 | 17.757 | 578 | 659 | IPR003035 | RWP-RK domain |
| Pfam | PF02042 | 7.3E-26 | 591 | 639 | IPR003035 | RWP-RK domain |
| SuperFamily | SSF54277 | 2.03E-22 | 861 | 948 | No hit | No description |
| SMART | SM00666 | 1.8E-28 | 863 | 945 | IPR000270 | PB1 domain |
| Gene3D | G3DSA:3.10.20.240 | 1.1E-26 | 863 | 945 | No hit | No description |
| PROSITE profile | PS51745 | 27.511 | 863 | 945 | IPR000270 | PB1 domain |
| Pfam | PF00564 | 4.1E-20 | 863 | 944 | IPR000270 | PB1 domain |
| CDD | cd06407 | 1.01E-33 | 864 | 944 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0010118 | Biological Process | stomatal movement | ||||
| GO:0010167 | Biological Process | response to nitrate | ||||
| GO:0042128 | Biological Process | nitrate assimilation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 960 aa Download sequence Send to blast |
MCEPDDHSAR NGVTTPPSRS RELLMDVDDL DLDGSWPLDQ IPYLSSSNRM ISPIFVSSSS 60 EQPCSPLWAF SDGGGNGFHH ATSGGDDEKI SSASGVPSFR LAEYPIFLPY SSPPAAENST 120 EKHNSFQFPS PLMSLVPPEN TDNYCVIKER MTQALRYFKE STEQNVLAQV WAPVRKNGRN 180 LLTTLGQPFV LNPNGNGLNQ YRMISLTYMF SVDSESDVEL GLPGRVFRQK LPEWTPNVQY 240 YSSKEFSRLD HALHYNVRGT LALPVFNPSG QSCIGVVELI MTSEKIHYAP EVDKVCKALE 300 AVNLKSSEIL DHQTTQICNE SRQNALAEIL EVLTVVCETH NLPLAQTWVP CQHGSVLANG 360 GGLKKNCTSF DGSCMGQICM STTDMACYVV DAHVWGFRDA CLEHHLQKGQ GVAGRAFLNG 420 GSCFCRDITK FCKTQYPLVH YALMFKLTTC FAISLQSSYT GDDSYILEFF LPSSITDDQE 480 QDSLLGSILV TMKEHFQSLR VASGVDFGED DDKLSFEIIQ ALPDKKVHSK IESIRVPFSG 540 FKSNAAETLL IPQPAVQSSD PVNEKINVSI VNGVVKEKKK TEKKRGKTEK TISLDVLQQY 600 FTGSLKDAAK SLGVCPTTMK RICRQHGISR WPSRKIKKVN RSITKLKRVI ESVQGTDGGL 660 DLTSMAVSSI PWTHGQTSAQ PLNSPNGSKP PELPNTNNSP NHWSSDHSPN EPNGSPELPP 720 SNGHKRSRTV DESAGTPTSH GSCDGNQLDE PKVPNQDSLF TVGGSPGLLF PPYSRDHDVT 780 AASFAMPNRL LGSIDHFRGM LIEDAGSSKD LRNLCSTSAF DDKFPDSNWM NNDNNINNNL 840 YAPPKEEAIA NVAREPSGLE MRTVTIKASY KEDIIRFRIS SGSGIMELKD EVAKRLKLDA 900 GTFDIKYLDD DNEWVLIACD ADLQECLEIP RSSRTKIVRL LIHDVTTNLG SSCESTGEL* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in regulation of nitrate assimilation and in transduction of the nitrate signal. {ECO:0000269|PubMed:18826430}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00449 | DAP | Transfer from AT4G24020 | Download |
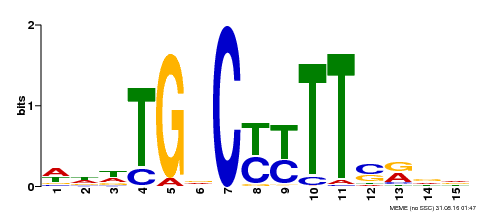
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.3358s0010.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Not regulated by the N source or by nitrate. {ECO:0000269|PubMed:18826430}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK228452 | 0.0 | AK228452.1 Arabidopsis thaliana mRNA for hypothetical protein, complete cds, clone: RAFL15-03-P13. | |||
| GenBank | BT005776 | 0.0 | BT005776.1 Arabidopsis thaliana clone RAFL15-03-P13 (R20174) unknown protein (At4g24020) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002869745.1 | 0.0 | protein NLP7 isoform X1 | ||||
| Swissprot | Q84TH9 | 0.0 | NLP7_ARATH; Protein NLP7 | ||||
| TrEMBL | D7M8Y6 | 0.0 | D7M8Y6_ARALL; RWP-RK domain-containing protein | ||||
| STRING | fgenesh2_kg.7__1893__AT4G24020.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2507 | 26 | 66 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G24020.1 | 0.0 | NIN like protein 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.3358s0010.1.p |



