 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.5127s0001.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 281aa MW: 31901.8 Da PI: 9.3714 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 86.5 | 2.7e-27 | 170 | 223 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k r++W+ eLH++Fv+av++L G++kA+Pk+ilelm+v+gL++e+v+SHLQk+R+
Araha.5127s0001.1.p 170 KSRVVWSIELHQQFVNAVNKL-GIDKAVPKRILELMNVPGLSRENVASHLQKFRM 223
68*******************.********************************8 PP
| |||||||
| 2 | Response_reg | 59.5 | 1.8e-20 | 7 | 94 | 23 | 109 |
CEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTTESEEEESS--HHHHHH CS
Response_reg 23 yeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaGakdflsKpfdpeelvk 109
y +v+ +++++ al++l+e++ +Dl+l D+ mpgm+G++ll++ e +lp+i+++ g +++ ++ Ga d+l Kp+ peel +
Araha.5127s0001.1.p 7 Y-QVTICSQADVALTILRERKdcFDLVLSDVHMPGMNGYKLLQQVGLLELDLPVIMMSVDGRTATVMTGINHGACDYLIKPIRPEELKN 94
6.89999**********99999****************************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 7.7E-127 | 1 | 278 | IPR017053 | Response regulator B-type, plant |
| CDD | cd00156 | 2.88E-21 | 1 | 99 | No hit | No description |
| SMART | SM00448 | 1.9E-10 | 1 | 96 | IPR001789 | Signal transduction response regulator, receiver domain |
| Gene3D | G3DSA:3.40.50.2300 | 9.7E-32 | 1 | 121 | No hit | No description |
| PROSITE profile | PS50110 | 34.032 | 1 | 100 | IPR001789 | Signal transduction response regulator, receiver domain |
| SuperFamily | SSF52172 | 2.13E-26 | 2 | 116 | IPR011006 | CheY-like superfamily |
| Pfam | PF00072 | 3.2E-17 | 7 | 94 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS51294 | 11.101 | 167 | 229 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.4E-28 | 167 | 228 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.11E-18 | 168 | 227 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.0E-24 | 170 | 223 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 7.3E-7 | 172 | 221 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 281 aa Download sequence Send to blast |
MLLRLMYQVT ICSQADVALT ILRERKDCFD LVLSDVHMPG MNGYKLLQQV GLLELDLPVI 60 MMSVDGRTAT VMTGINHGAC DYLIKPIRPE ELKNIWQHVV RRKCVMKKEL RNSQALEDNK 120 NSGSLETVFS ASECSEGSLM KRRKKKKKRS VDREDNEDDL DLLNPGNSKK SRVVWSIELH 180 QQFVNAVNKL GIDKAVPKRI LELMNVPGLS RENVASHLQK FRMYLKRLSG EASQNNDTES 240 IKRYENIQAL VSSGQLHPQT LAALYGQTID NHHSASFGVW I |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 1e-20 | 169 | 229 | 4 | 64 | ARR10-B |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 138 | 146 | LMKRRKKKK |
| 2 | 140 | 148 | KRRKKKKKR |
| 3 | 141 | 147 | RRKKKKK |
| 4 | 142 | 148 | RKKKKKR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
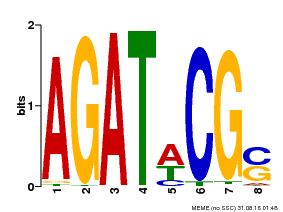
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-[AG]GATT-3'. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. Could directly activate some type-A response regulators in response to cytokinins (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00098 | PBM | Transfer from AT2G01760 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.5127s0001.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY088149 | 0.0 | AY088149.1 Arabidopsis thaliana clone 41875 mRNA, complete sequence. | |||
| GenBank | AY099726 | 0.0 | AY099726.1 Arabidopsis thaliana putative two-component response regulator protein (At2g01760) mRNA, complete cds. | |||
| GenBank | AY128887 | 0.0 | AY128887.1 Arabidopsis thaliana putative two-component response regulator protein (At2g01760) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002875093.1 | 0.0 | two-component response regulator ARR14 isoform X2 | ||||
| Swissprot | Q8L9Y3 | 1e-176 | ARR14_ARATH; Two-component response regulator ARR14 | ||||
| TrEMBL | D7LPV7 | 0.0 | D7LPV7_ARALL; Two-component response regulator | ||||
| STRING | scaffold_500093.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM8150 | 27 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01760.1 | 1e-165 | response regulator 14 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.5127s0001.1.p |



