 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.60889s0001.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 566aa MW: 63986.5 Da PI: 5.263 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 433.6 | 1.8e-132 | 38 | 401 | 1 | 351 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasd 89
e+l++rmwkd+++lkr+ker+k + a +++ +k+++qa+rkkmsraQDgiLkYMlk mevc+++GfvYgiipekgkpv+g+sd
Araha.60889s0001.1.p 38 EDLERRMWKDRVRLKRIKERQKVGS--QGA-QTKETPKKISDQAQRKKMSRAQDGILKYMLKLMEVCKVRGFVYGIIPEKGKPVSGSSD 123
89*****************999754..444.5677899*************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S-- CS
EIN3 90 sLraWWkekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGke 178
++raWWkekv+fd+ngpaai+ky+ ++l++++++++ ++++ l++lqD+tlgSLLs+lmqhcdppqr++plekg++pPWWPtG+e
Araha.60889s0001.1.p 124 NIRAWWKEKVKFDKNGPAAIAKYEEECLAFGKSDGN----RNSQFVLQDLQDATLGSLLSSLMQHCDPPQRKYPLEKGTPPPWWPTGNE 208
******************************987777....77999******************************************** PP
HHHHHHT--TT--.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX..XX CS
EIN3 179 lwwgelglskdqgtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah..ss 265
+ww +lgl+k+q+ ppy+kphdlkk+wkv+vLtavi+hm p+i++i++++rqsk+lqdkm+akes+++l+vlnqee+++++ s++ +s
Araha.60889s0001.1.p 209 EWWVKLGLPKSQS-PPYRKPHDLKKMWKVGVLTAVINHMLPDIAKIKRHVRQSKCLQDKMTAKESAIWLAVLNQEESLIQQPSSDngNS 296
*************.9**********************************************************************6522 PP
........XXXXXXXXXXXXXXXXXXXXXXXXXX.XXXXXXXXXX..................XXXXXXXXXXXXXXXXXXXXX..... CS
EIN3 266 ........slrkqspkvtlsceqkedvegkkeskikhvqavktta..................gfpvvrkrkkkpsesakvsskevsrt 328
+ +++k ++++++++dv+g+++ + + ++++++++ +++ +r+rk+++ + + + +
Araha.60889s0001.1.p 297 nvteahrrGNNADRRKPVINSDSDYDVDGTEDASGSVSSKDSRRNqtqkeqptaishivrdqdKAEKHRRRKRPRIR-------SGTNN 378
67876544333378889999999*****6666666666666665566777777787666543333333333322222.......22355 PP
.XXXXXXX.XXXXXXXXXXXXXX CS
EIN3 329 cqssqfrgsetelifadknsisq 351
q+++ ++ e+++i++d+n++++
Araha.60889s0001.1.p 379 RQEEEQPEAEQRNILPDMNHVDD 401
67778888999*******99876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 3.2E-126 | 38 | 283 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 7.6E-71 | 159 | 291 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 3.27E-60 | 164 | 286 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 566 aa Download sequence Send to blast |
MGDLAGSVAD IRMENEPDDL ASDNVAEIDV SDEEIDAEDL ERRMWKDRVR LKRIKERQKV 60 GSQGAQTKET PKKISDQAQR KKMSRAQDGI LKYMLKLMEV CKVRGFVYGI IPEKGKPVSG 120 SSDNIRAWWK EKVKFDKNGP AAIAKYEEEC LAFGKSDGNR NSQFVLQDLQ DATLGSLLSS 180 LMQHCDPPQR KYPLEKGTPP PWWPTGNEEW WVKLGLPKSQ SPPYRKPHDL KKMWKVGVLT 240 AVINHMLPDI AKIKRHVRQS KCLQDKMTAK ESAIWLAVLN QEESLIQQPS SDNGNSNVTE 300 AHRRGNNADR RKPVINSDSD YDVDGTEDAS GSVSSKDSRR NQTQKEQPTA ISHIVRDQDK 360 AEKHRRRKRP RIRSGTNNRQ EEEQPEAEQR NILPDMNHVD DPLLEYNING THHEEDVVDP 420 NIALGPEENG LELVVPEFHN NYTYLPPVNR QDMMPVDERP MLYGPNPNHE LQFGSGYNFY 480 NPSAVFVHNQ EDDILHTQIE MNAQAPPHNS GFEAPGGVLQ PLGLLGNEDG LAGSELPPEY 540 PSDIMSPFND LSFDYGIDDF SWFGA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 5e-87 | 163 | 294 | 9 | 140 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
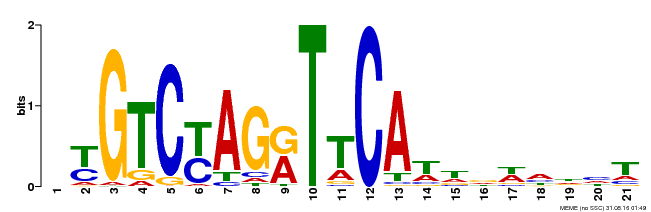
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.60889s0001.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF004215 | 0.0 | AF004215.1 Arabidopsis thaliana ethylene-insensitive3-like3 (EIL3) mRNA, complete cds. | |||
| GenBank | AY070044 | 0.0 | AY070044.1 Arabidopsis thaliana putative ethylene-insensitive protein EIL3 (At1g73730) mRNA, complete cds. | |||
| GenBank | AY133839 | 0.0 | AY133839.1 Arabidopsis thaliana clone U18784 putative ethylene-insensitive protein EIL3 (At1g73730) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002888943.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 0.0 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | D7KRL6 | 0.0 | D7KRL6_ARALL; Uncharacterized protein | ||||
| STRING | scaffold_202309.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4658 | 25 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 0.0 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.60889s0001.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



