 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.6714s0002.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 787aa MW: 87630.4 Da PI: 5.3007 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 135.2 | 2e-42 | 121 | 198 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqve+Ceadls++k+yhrrhkvCe+hska++++v g+ qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
Araha.6714s0002.1.p 121 VCQVENCEADLSKVKDYHRRHKVCEMHSKATSAIVGGILQRFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTNP 198
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51141 | 32.787 | 119 | 196 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:4.10.1100.10 | 1.5E-33 | 119 | 183 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.31E-39 | 120 | 200 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 5.9E-31 | 122 | 195 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 3.4E-7 | 689 | 784 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 4.35E-7 | 693 | 785 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 8.667 | 695 | 787 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.13E-8 | 695 | 785 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 787 aa Download sequence Send to blast |
MEGEEAQHFY GFRAVDLRSV GKTSVEWDLN DWKWDGDLFL ATQLNPAASE TTGRQLFPLG 60 NSSNSSSSCS DEGNNTNMVE KKRRAVAIQG DSNAAGGLTL KLGDNACHLN AAKKTKFGAA 120 VCQVENCEAD LSKVKDYHRR HKVCEMHSKA TSAIVGGILQ RFCQQCSRFH LLQEFDEGKR 180 SCRRRLAGHN KRRRKTNPEP GANGNPSDDQ SSNYLLISLL KILSNMHNHT GDQDLMSHLL 240 KSLVSHAGEQ LGKNLVELLL QGGSQGSLNI GNSALLGIEQ APQEDLKQLS VNVPRQDGTA 300 TETRSEKQVK MNDFDLNDIY IDSDDTDIER SPPPTNPATS SLDYPSWIHQ SSPPQTSRNS 360 DSASDQSPSS SSEDAQMRTG RIVFKLFGKE PNEFPIVLRG QILDWLSHSP TDMESYIRPG 420 CIVLTIYLRQ AETAWEELLD DLGFSLRKLL DLSDDPLWTT GWIYVRVQNQ LAFVYNGQVV 480 VDTSLPLKSL DYSHIISVKP LAIAATGKAQ FTVKGINLRR RGTRLLCAVE GKYLIQETTH 540 DSTTGENDDL KENNEIVECV NFSCDMPITS GRGYMEIEDQ GLSSSFFPFL VVEDDDVCSE 600 IRILETTLEF TGTDSAKQAM DFIHEIGWLL HRSKLGESDP NPDVFPLIRF QWLIEFSMDR 660 EWCAVIRKLL NMFFDGAVGE FSSSSNATLS ELCLLHRAVR KNSKPMVEML LRYVPKKQRN 720 SLFRPDAAGP AGLTPLHIAA GKDGSEDVLD ALTEDPAMVG IEAWKTSRDS TGFTPEDYAR 780 LRGHFSY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 3e-32 | 118 | 195 | 7 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
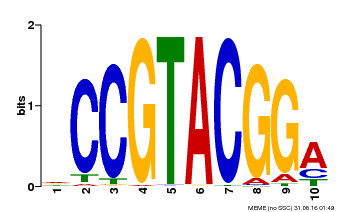
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' of AP1 promoter. Binds specifically to the 5'-GTAC-3' core sequence. {ECO:0000269|PubMed:10524240, ECO:0000269|PubMed:16095614}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00603 | PBM | Transfer from AT2G47070 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.6714s0002.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AJ011629 | 0.0 | AJ011629.1 Arabidopsis thaliana (ecotype Columbia) mRNA for squamosa promoter binding protein-like 1. | |||
| GenBank | AK230269 | 0.0 | AK230269.1 Arabidopsis thaliana mRNA for squamosa promoter binding protein-like 1, complete cds, clone: RAFL23-28-L05. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020883222.1 | 0.0 | squamosa promoter-binding-like protein 1 | ||||
| Swissprot | Q9SMX9 | 0.0 | SPL1_ARATH; Squamosa promoter-binding-like protein 1 | ||||
| TrEMBL | D7LFY4 | 0.0 | D7LFY4_ARALL; Uncharacterized protein | ||||
| STRING | fgenesh2_kg.4__2943__AT2G47070.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3030 | 27 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.6714s0002.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



