 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.6872s0001.1.p | ||||||||
| Common Name | 8F2.12 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 341aa MW: 37405.1 Da PI: 6.0805 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 59.8 | 6.3e-19 | 118 | 168 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
++++GVr+++ +g+W+AeIrdp + r r +lg++ taeeAa +++a+ +l+g
Araha.6872s0001.1.p 118 KKFRGVRQRP-WGKWAAEIRDPLK---RVRLWLGTYNTAEEAAMVYDNAAIQLRG 168
59********.**********754...5*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 2.24E-32 | 118 | 175 | No hit | No description |
| Pfam | PF00847 | 6.7E-12 | 119 | 168 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.0E-29 | 119 | 176 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 23.406 | 119 | 176 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.05E-21 | 119 | 177 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 3.3E-37 | 119 | 182 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 7.6E-10 | 120 | 131 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 7.6E-10 | 142 | 158 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042991 | Biological Process | transcription factor import into nucleus | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048825 | Biological Process | cotyledon development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 341 aa Download sequence Send to blast |
MEAEKKMVLP RIKFTEHKTN TTTIVPDLTN NHQTRVLRIS VTDPDATDSS SDDEEEEHQR 60 FVSKRRRIKK FVNEVVLDSG AVAGSCSQME SKKRGKSAVK SESVVSPVVS ATTTTTEKKF 120 RGVRQRPWGK WAAEIRDPLK RVRLWLGTYN TAEEAAMVYD NAAIQLRGPD ALTNFSVTPT 180 TTEKKPPPPP SPVKKKKKTS KVKKSVTASS SISRSSSNDC LCSPVSVLRS PFAVDEFSGI 240 SSPPPAAVVV KEEPSMTTVS ETFSDFSAPL FSDDDVFDFR SSVVPDYLGG DLFGEDLFTA 300 DMCTDMNFGF DFGSGLSSWH LEDHFQDIGD LFGSDPLLAV * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 1e-20 | 118 | 183 | 4 | 70 | ATERF1 |
| 3gcc_A | 1e-20 | 118 | 183 | 4 | 70 | ATERF1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 63 | 67 | KRRRI |
| 2 | 193 | 203 | KKKKKTSKVKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Component of the cytokinin signaling pathway involved in cotyledons, leaves, and embryos development. Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250, ECO:0000269|PubMed:16832061}. | |||||
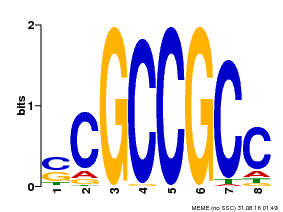
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00027 | PBM | Transfer from AT4G23750 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.6872s0001.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cytokinins. {ECO:0000269|PubMed:16832061}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EF197846 | 0.0 | EF197846.1 Arabidopsis halleri miR319a microRNA locus, genomic sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_194106.1 | 0.0 | cytokinin response factor 2 | ||||
| Refseq | NP_974599.1 | 0.0 | cytokinin response factor 2 | ||||
| Refseq | XP_020873605.1 | 0.0 | ethylene-responsive transcription factor CRF2 | ||||
| Swissprot | Q9SUQ2 | 0.0 | CRF2_ARATH; Ethylene-responsive transcription factor CRF2 | ||||
| TrEMBL | B2WS71 | 0.0 | B2WS71_ARAHA; Uncharacterized protein | ||||
| STRING | AT4G23750.1 | 0.0 | (Arabidopsis thaliana) | ||||
| STRING | fgenesh2_kg.7__1924__AT4G23750.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10 | 28 | 1650 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G23750.2 | 1e-155 | cytokinin response factor 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.6872s0001.1.p |



