 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.7279s0004.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 193aa MW: 21819.2 Da PI: 10.8517 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 45 | 2.3e-14 | 112 | 162 | 5 | 54 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH.HHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkke.leelkk 54
+r++r++kNRe+A rsR+RK+a++ eLe ++++L++ N++L+ + +e ++k
Araha.7279s0004.1.p 112 RRQKRMIKNRESAARSRARKQAYTMELEAEIAQLKELNEELQRKqVEIMEK 162
69*************************************996540444444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 1.1E-13 | 108 | 173 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.45 | 110 | 155 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14707 | 1.72E-23 | 112 | 166 | No hit | No description |
| SuperFamily | SSF57959 | 2.25E-10 | 112 | 165 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 8.6E-14 | 112 | 168 | No hit | No description |
| Pfam | PF00170 | 4.0E-12 | 112 | 169 | IPR004827 | Basic-leucine zipper domain |
| PROSITE pattern | PS00036 | 0 | 115 | 130 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 193 aa Download sequence Send to blast |
QQLIQKQERP FPKQTTIAFS NTADVVNRSQ PATQCQEVKP SILGIRNHPV NNNLLQAVDF 60 KTGVTVAAVS PGSQMSPDLT PKSALDASLS PVPYMFGRVR KTGAVLEKVI ERRQKRMIKN 120 RESAARSRAR KQAYTMELEA EIAQLKELNE ELQRKQVEIM EKQKNQLLEP LRQPWGMGCK 180 RQCLRRTLTG PW* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the ABA-responsive element (ABRE). Mediates stress-responsive ABA signaling. {ECO:0000269|PubMed:11884679, ECO:0000269|PubMed:15361142}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00038 | PBM | Transfer from AT4G34000 | Download |
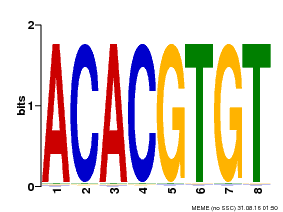
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.7279s0004.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by drought, salt, abscisic acid (ABA). {ECO:0000269|PubMed:10636868, ECO:0000269|PubMed:16284313}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF093546 | 0.0 | AF093546.1 Arabidopsis thaliana clone 11 abscisic acid responsive elements-binding factor (ABRE) mRNA, complete cds. | |||
| GenBank | AK175851 | 0.0 | AK175851.1 Arabidopsis thaliana mRNA for abscisic acid responsive elements-binding factor (ABRE/ABF3), complete cds, clone: RAFL22-47-B14. | |||
| GenBank | AY054605 | 0.0 | AY054605.1 Arabidopsis thaliana Unknown protein (F17I5.190:F17I5.200) mRNA, complete cds. | |||
| GenBank | AY081467 | 0.0 | AY081467.1 Arabidopsis thaliana unknown protein (F17I5.190:F17I5.200) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001320130.1 | 1e-130 | abscisic acid responsive elements-binding factor 3 | ||||
| Refseq | NP_567949.1 | 1e-130 | abscisic acid responsive elements-binding factor 3 | ||||
| Refseq | NP_849490.2 | 1e-130 | abscisic acid responsive elements-binding factor 3 | ||||
| Swissprot | Q9M7Q3 | 1e-131 | AI5L6_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 6 | ||||
| TrEMBL | D7MFS7 | 1e-127 | D7MFS7_ARALL; Uncharacterized protein | ||||
| STRING | AT4G34000.1 | 1e-129 | (Arabidopsis thaliana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM734 | 27 | 129 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G34000.2 | 1e-133 | abscisic acid responsive elements-binding factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.7279s0004.1.p |



