 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.7659s0001.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 413aa MW: 46195.1 Da PI: 4.8051 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 55.8 | 1.1e-17 | 22 | 69 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+ Ed ll+ +v ++G g+W+++ ++ g+ R++k+c++rw ++l
Araha.7659s0001.1.p 22 KGPWTQAEDNLLIAYVDKHGDGNWNAVQKHSGLSRCGKSCRLRWVNHL 69
79*******************************************996 PP
| |||||||
| 2 | Myb_DNA-binding | 46.5 | 8.2e-15 | 75 | 118 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g++T+ E+++ ++++++lG++ W++ a +++ gRt++++k++w++
Araha.7659s0001.1.p 75 KGAFTEKEEKRVIELHALLGNK-WARMAGELP-GRTDNEIKNFWNT 118
799*******************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.259 | 17 | 69 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.12E-28 | 20 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.3E-15 | 21 | 71 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.7E-16 | 22 | 69 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-23 | 23 | 76 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.05E-12 | 24 | 69 | No hit | No description |
| PROSITE profile | PS51294 | 23.817 | 70 | 124 | IPR017930 | Myb domain |
| SMART | SM00717 | 7.9E-14 | 74 | 122 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.0E-13 | 75 | 118 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 7.03E-10 | 77 | 120 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-23 | 77 | 122 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 413 aa Download sequence Send to blast |
MGKVRHDSGS DDETSIDQSF TKGPWTQAED NLLIAYVDKH GDGNWNAVQK HSGLSRCGKS 60 CRLRWVNHLR PDLKKGAFTE KEEKRVIELH ALLGNKWARM AGELPGRTDN EIKNFWNTRL 120 KRLQRLGLPV YPDEVREQAM NAAIQSGLNT DSLDGHHSQD SLEADCVEIP ELNFQHLPLN 180 QCSSYYQSML RNVPPTGVFV RQRPCFFQPN MYNLITTPPY MSTGKRSREP ETAFPGGYVM 240 NEQSPPLWNY PFVENISEQL PDSHLLGNAT YSSSSPGPLI HGVEKLELPS FQYLEEPGGW 300 GAEQSDQMPE HHESDNTLVQ SPLTDQTPSD CPSSLYDGLL ESVVYGSSGE KPATDTDSES 360 SLFQSSLLGH TEFTPANENR TDLGSTSSNE GRITFEGDWI RLLLGEDRYS TK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 7e-30 | 20 | 122 | 5 | 106 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00279 | DAP | Transfer from AT2G26960 | Download |
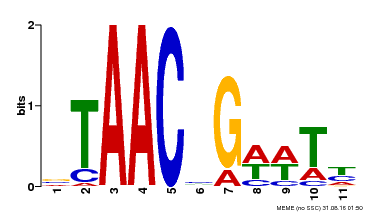
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.7659s0001.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ446571 | 0.0 | DQ446571.1 Arabidopsis thaliana clone pENTR221-At2g26960 myb family transcription factor (At2g26960) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002880852.2 | 0.0 | transcription factor MYB104 | ||||
| Swissprot | Q9SM27 | 1e-130 | MY104_ARATH; Transcription factor MYB104 | ||||
| TrEMBL | D7LFF1 | 0.0 | D7LFF1_ARALL; Predicted protein | ||||
| STRING | Al_scaffold_0004_975 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM9198 | 21 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G26960.1 | 0.0 | myb domain protein 81 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.7659s0001.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



