 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araha.8818s0009.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 464aa MW: 50241.7 Da PI: 5.2767 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 124 | 4.8e-39 | 140 | 199 | 2 | 61 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkks 61
++k+l+cprC s++tkfCyynny+++qPr+fCk+C+ryWt+GG++rnvPvG+grrknk+
Araha.8818s0009.1.p 140 PDKILPCPRCSSMETKFCYYNNYNVNQPRHFCKKCQRYWTAGGTMRNVPVGAGRRKNKSP 199
7899*****************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 3.0E-31 | 136 | 197 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 8.2E-32 | 142 | 198 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 28.444 | 144 | 198 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 146 | 182 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 464 aa Download sequence Send to blast |
MADPAIKLFG KTIPLPELAV VVDSCSSYTG VLTETQDQNL VRLSDSCIGD DDDEMGRSGL 60 GGGDDDGGDG SRVGGESESD KKEEKNNECQ EESLRHESND VTSGITEKTE TTKAAKTNEE 120 SSQNETGGTA CSQEAKLKKP DKILPCPRCS SMETKFCYYN NYNVNQPRHF CKKCQRYWTA 180 GGTMRNVPVG AGRRKNKSPA SHYNRHVSIT SAEAMQKAAR TDLQHPNGAN LLTFGSDSVL 240 CESMASGLNL AEKSMLKTQT VLQEPNEGLK ITVPLNPSNE EAGTVSPLPK VPCFPGPPPA 300 WPYAWNGVSW TVLPFYPPPA YWSCPGVSPG TWNSFTWMPQ PNSPSGSGPN SPTLGKHSRD 360 ENAAEPGTTF EETESLGREK SKPERCLWVP KTLRVDDPEE AAKSSIWETL GIKKDENADT 420 FGAFRSPTKE KSSLSEGKLP GRRAELQANP AALSRSANFH ESS* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
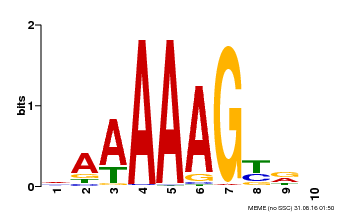
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence (By similarity). Regulates a photoperiodic flowering response. Transcriptional repressor of 'CONSTANS' expression. The stability of CDF2 is controlled by 'GIGANTEA' and redundantly by ADO3, ADO2 and/or ADO1. {ECO:0000250, ECO:0000269|PubMed:19619493}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00046 | PBM | Transfer from AT5G39660 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araha.8818s0009.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian-regulation. Highly expressed at the beginning of the light period, then decreases, reaching a minimum between 16 and 29 hours after dawn before rising again at the end of the day. Regulated at the protein level by ADO3 and GI pos-transcriptionally. {ECO:0000269|PubMed:19619493}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT015791 | 0.0 | BT015791.1 Arabidopsis thaliana At5g39660 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002870770.1 | 0.0 | cyclic dof factor 2 | ||||
| Swissprot | Q93ZL5 | 0.0 | CDF2_ARATH; Cyclic dof factor 2 | ||||
| TrEMBL | D7MJJ7 | 0.0 | D7MJJ7_ARALL; Uncharacterized protein | ||||
| STRING | scaffold_704079.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13795 | 18 | 25 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G39660.2 | 0.0 | cycling DOF factor 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Araha.8818s0009.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



