 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.2L6E3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 262aa MW: 29294 Da PI: 8.0642 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 54.4 | 2.8e-17 | 14 | 62 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT.-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGgg.tWktIartmgkgRtlkqcksrwqkyl 48
+g+W++eEd +l+ +++q+G+g +W + ++++g++R++k+c++rw++yl
Araip.2L6E3 14 KGPWSPEEDAKLKSYIEQHGTGgNWIALPQKIGLKRCGKSCRLRWLNYL 62
79*********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 38.4 | 2.8e-12 | 69 | 111 | 2 | 46 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
g++++eEd ++ + G++ W+ Ia+ ++ gRt++++k++w++
Araip.2L6E3 69 GSFSEEEDNIICTLYVSIGSR-WSIIAAQLP-GRTDNDIKNYWNT 111
78*******************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.467 | 9 | 62 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 6.27E-27 | 11 | 109 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.4E-13 | 13 | 64 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.0E-16 | 14 | 62 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-25 | 15 | 69 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.48E-10 | 16 | 62 | No hit | No description |
| PROSITE profile | PS51294 | 22.679 | 63 | 117 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.4E-11 | 67 | 115 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.5E-11 | 69 | 111 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.9E-24 | 70 | 117 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.51E-8 | 70 | 113 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 262 aa Download sequence Send to blast |
MGRAPCCDKA NVKKGPWSPE EDAKLKSYIE QHGTGGNWIA LPQKIGLKRC GKSCRLRWLN 60 YLRPNIKHGS FSEEEDNIIC TLYVSIGSRW SIIAAQLPGR TDNDIKNYWN TRLKKKLLAK 120 QRKLDQPPPI PYCNSSSNSS SSSSSVPQLV VPPAPPSIHD YDFTSLISSF PQQHHHLYPP 180 ANMYHHQQGF ESLIDDDLSH LMACVVSNEQ GFYGSTTSTE STTTSWGPDM TSLVNYSSSP 240 PPPLLYQDDA SPASRYFGMQ ML |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 9e-26 | 14 | 117 | 7 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00089 | SELEX | Transfer from AT3G49690 | Download |
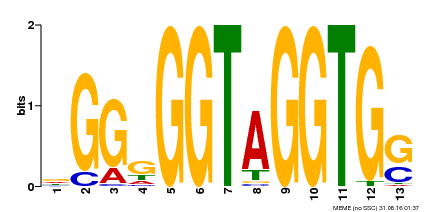
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.2L6E3 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT139688 | 1e-105 | BT139688.1 Lotus japonicus clone JCVI-FLLj-19M6 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016201583.1 | 0.0 | transcription factor MYB36 | ||||
| TrEMBL | A0A444WUE7 | 1e-159 | A0A444WUE7_ARAHY; Uncharacterized protein | ||||
| STRING | AES94491 | 2e-86 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF130 | 34 | 340 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G49690.1 | 2e-76 | myb domain protein 84 | ||||



