 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.98LTB | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 283aa MW: 31307.9 Da PI: 10.2823 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 356.6 | 5.1e-109 | 1 | 283 | 1 | 301 |
GAGA_bind 1 mdddgsre.rnkgyyepaaslkenlglqlmssiaerdaki.rernlalsekkaavaerd.........maflqrdkalaernkalverdnkllalllv 87
md d+ + rn+gyyepa+s+k++lglqlmss++e+ r++ + ls++++a+++rd m++ +rd+++++ +++k+++++++
Araip.98LTB 1 MDGDNGLNiRNWGYYEPATSFKSHLGLQLMSSMPEKPLLGgRNAAV-LSGTNGAYHHRDigmppatypMDY-MRDAWISS-------QREKYMNMIPT 89
888888888*************************998766455555.9***********************.********.......889*******6 PP
GAGA_bind 88 enslasalpvgvqvlsgtksidslqqlsepqledsavelreeeklealpieeaaeeakekkkkkkrqrakkpkekkakkkkkksekskkkvkkesade 185
+ +++++++t+s++++q+ ++ +e + +e+ p+eea+ ++k +++ kkrq +k+pk++kakk+k+ ++++k+++++ +
Araip.98LTB 90 NP--------SYGSIPETSSTHHMQM----------IPPPELSAKEERPVEEAPVVEKANTTGKKRQGPKVPKSPKAKKPKRGPRVPKDENAPTV--Q 167
55........599*************..........23345566677888888888889999999999**************9999977777776..7 PP
GAGA_bind 186 rskaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSvyPLPvstkrrgaRiagrKmSqgafkklLekLaaeGydlsnpv 283
r++a+kk++++v+ng++lD+s++P+PvCsCtG+++qCY+WG+GGWqSaCCtt +S+yPLP+stkrrgaRiagrKmS gafkk+LekLaaeGy++snp+
Araip.98LTB 168 RARAPKKTTEIVINGIDLDISSIPIPVCSCTGTPQQCYRWGSGGWQSACCTTGMSTYPLPMSTKRRGARIAGRKMSIGAFKKVLEKLAAEGYNFSNPI 265
9************************************************************************************************* PP
GAGA_bind 284 DLkdhWAkHGtnkfvtir 301
DL+++WAkHGtnkfvtir
Araip.98LTB 266 DLRTYWAKHGTNKFVTIR 283
*****************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF06217 | 3.7E-113 | 1 | 283 | IPR010409 | GAGA-binding transcriptional activator |
| SMART | SM01226 | 1.0E-169 | 1 | 283 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0050793 | Biological Process | regulation of developmental process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 283 aa Download sequence Send to blast |
MDGDNGLNIR NWGYYEPATS FKSHLGLQLM SSMPEKPLLG GRNAAVLSGT NGAYHHRDIG 60 MPPATYPMDY MRDAWISSQR EKYMNMIPTN PSYGSIPETS STHHMQMIPP PELSAKEERP 120 VEEAPVVEKA NTTGKKRQGP KVPKSPKAKK PKRGPRVPKD ENAPTVQRAR APKKTTEIVI 180 NGIDLDISSI PIPVCSCTGT PQQCYRWGSG GWQSACCTTG MSTYPLPMST KRRGARIAGR 240 KMSIGAFKKV LEKLAAEGYN FSNPIDLRTY WAKHGTNKFV TIR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. Negatively regulates the homeotic gene AGL11/STK, which controls ovule primordium identity, by a cooperative binding to purine-rich elements present in the regulatory sequence leading to DNA conformational changes. {ECO:0000269|PubMed:14731261, ECO:0000269|PubMed:15722463}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00253 | DAP | Transfer from AT2G01930 | Download |
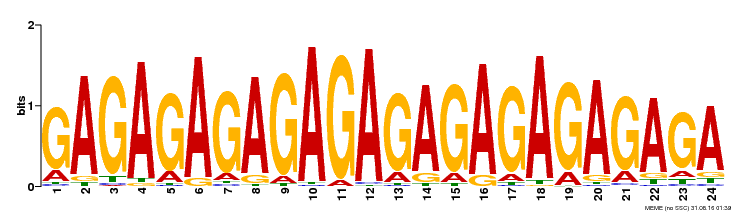
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.98LTB |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015966841.1 | 0.0 | protein BASIC PENTACYSTEINE2 | ||||
| Refseq | XP_015966842.1 | 0.0 | protein BASIC PENTACYSTEINE2 | ||||
| Refseq | XP_016203362.1 | 0.0 | protein BASIC PENTACYSTEINE2 | ||||
| Refseq | XP_016203363.1 | 0.0 | protein BASIC PENTACYSTEINE2 | ||||
| Refseq | XP_020958387.1 | 0.0 | protein BASIC PENTACYSTEINE2 | ||||
| Refseq | XP_025655545.1 | 0.0 | protein BASIC PENTACYSTEINE2 isoform X1 | ||||
| Refseq | XP_025655546.1 | 0.0 | protein BASIC PENTACYSTEINE2 isoform X1 | ||||
| Swissprot | Q9SKD0 | 1e-96 | BPC1_ARATH; Protein BASIC PENTACYSTEINE1 | ||||
| TrEMBL | A0A445D1C4 | 0.0 | A0A445D1C4_ARAHY; Uncharacterized protein | ||||
| STRING | GLYMA09G30040.4 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2370 | 32 | 83 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G14685.3 | 5e-90 | basic pentacysteine 2 | ||||



