 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.9P5VU | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 332aa MW: 37746.3 Da PI: 7.1454 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 24.2 | 7.9e-08 | 43 | 88 | 3 | 46 |
SS-HHHHHHHHHHHHHTTTT...-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGgg...tWktIartmgkgRtlkqcksrwqk 46
+WT++E +++ +a +++ ++ +W ++a++++ g+t +++ ++
Araip.9P5VU 43 KWTPQENKQFENALALFDKEtpdRWLKVAAMIP-GKTVGDVIKQYRE 88
8*****************99*************.*******999986 PP
| |||||||
| 2 | Myb_DNA-binding | 47.4 | 4.4e-15 | 151 | 195 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ l++ + k++G+g+W+ I+r + ++Rt+ q+ s+ qky
Araip.9P5VU 151 PWTEEEHRLFLMGLKKYGKGDWRNISRNFVTTRTPTQVASHAQKY 195
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 10.182 | 39 | 94 | IPR017884 | SANT domain |
| SMART | SM00717 | 4.8E-9 | 40 | 92 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.79E-11 | 42 | 98 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.30E-7 | 43 | 90 | No hit | No description |
| Pfam | PF00249 | 1.2E-6 | 43 | 89 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.2E-13 | 142 | 201 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 20.858 | 144 | 200 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.73E-17 | 145 | 199 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.4E-17 | 147 | 199 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 1.7E-13 | 148 | 198 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.20E-11 | 151 | 196 | No hit | No description |
| Pfam | PF00249 | 1.8E-12 | 151 | 195 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 332 aa Download sequence Send to blast |
MYATIAYSLL EEEEEMNRGI EVLSPASYLH NSNWLFQESA GTKWTPQENK QFENALALFD 60 KETPDRWLKV AAMIPGKTVG DVIKQYRELV EDVNVIESGF IPIPGGTTDG SFTLEWVDNH 120 EFDGFKQFYG VGCKRGSSTR PSEQERKKGV PWTEEEHRLF LMGLKKYGKG DWRNISRNFV 180 TTRTPTQVAS HAQKYFIRQL NGGKDKRRSS IHDITMVNLP ETKSPSSESN KPPSPDANLN 240 QNQKLLSSMV KQEYDWKSPP YEGVPLLFDS TNGNSFMTPL CGASSYGSKP QEQYLLRGTF 300 HGFQFGPYDT FMQMQPQKDL SGRMSRAQGS YV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 6e-16 | 40 | 110 | 7 | 77 | RADIALIS |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the dorsovental asymmetry of flowers. Promotes ventral identity. {ECO:0000269|PubMed:11937495, ECO:0000269|PubMed:9118809}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00296 | DAP | Transfer from AT2G38090 | Download |
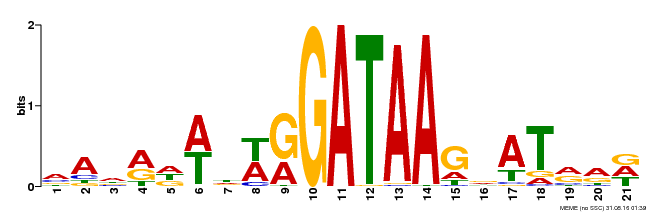
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.9P5VU |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016183904.1 | 0.0 | transcription factor DIVARICATA | ||||
| Refseq | XP_025633695.1 | 0.0 | transcription factor DIVARICATA | ||||
| Swissprot | Q8S9H7 | 1e-117 | DIV_ANTMA; Transcription factor DIVARICATA | ||||
| TrEMBL | A0A445AK09 | 0.0 | A0A445AK09_ARAHY; Uncharacterized protein | ||||
| STRING | GLYMA03G14440.1 | 1e-171 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF466 | 34 | 162 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38090.1 | 1e-112 | MYB family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



