 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.CZ9NC | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 320aa MW: 35402.4 Da PI: 7.8812 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 42.9 | 1.1e-13 | 124 | 168 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ ++ + ++lG+g+W+ I+r ++Rt+ q+ s+ qky
Araip.CZ9NC 124 PWTEEEHRTFLVGLEKLGKGDWRGISRNYVTTRTPTQVASHAQKY 168
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50158 | 8.532 | 3 | 18 | IPR001878 | Zinc finger, CCHC-type |
| PROSITE profile | PS51294 | 17.764 | 117 | 173 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.79E-17 | 119 | 173 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.3E-18 | 120 | 171 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 2.2E-10 | 121 | 171 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.6E-11 | 121 | 167 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 2.2E-11 | 124 | 168 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.96E-9 | 124 | 169 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 320 aa Download sequence Send to blast |
MGRKCSHCGN IGHNSRTCTT SPMMMMMRGG TNNNNNNNSF VGLRLFGVQL DISSSSCCVA 60 MKKSFSMDSL PSSSSTTTTT TSSFSSSRLP IDDNITLNNN NSSAFGYLSD GLIVPPQDRK 120 KGVPWTEEEH RTFLVGLEKL GKGDWRGISR NYVTTRTPTQ VASHAQKYFL RLATINKKKR 180 RSSLFDLVGI GNNNTKTEFH DDVGGISNCK SGDSSVIVCN ESKLEEVVEN NDIDHATLSL 240 LGRLTPFHQE IKSDNDLKVN TINNDNYNHN HQPFWPHHHH QQHMNSSSTV PDLELTLAAP 300 EDQNKSTSPS SFFFGPISVT |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 176 | 180 | KKKRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00565 | DAP | Transfer from AT5G56840 | Download |
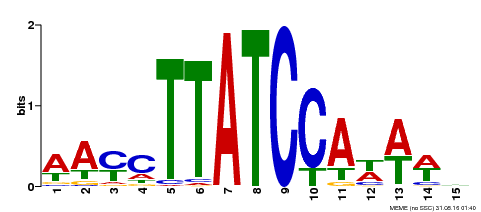
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.CZ9NC |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF208672 | 0.0 | KF208672.1 Arachis hypogaea putative MYB-related protein 18 (MYB18) gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016200943.1 | 0.0 | myb-like protein I | ||||
| Refseq | XP_025656218.1 | 0.0 | myb-like protein I | ||||
| TrEMBL | A0A445D3T2 | 0.0 | A0A445D3T2_ARAHY; Uncharacterized protein | ||||
| STRING | XP_004513098.1 | 8e-97 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3213 | 32 | 71 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56840.1 | 2e-57 | MYB_related family protein | ||||



