 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.EDM7N | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 256aa MW: 29056.8 Da PI: 9.9197 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 92.5 | 8.5e-29 | 56 | 170 | 3 | 89 |
TCP 3 gkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgt..................................s 66
g+kdrhsk++T +g+RdRR+Rls+++a++++dLqd+LG+ ++sk i+WLl + k i++l+ + s
Araip.EDM7N 56 GGKDRHSKVCTIRGLRDRRIRLSVPTAIQLYDLQDKLGLSQPSKVIDWLLDATKLDIDKLPPLhfpqpcfqppqtlfphllhdpssfwkprfweldsS 153
79*************************************************************88888777777777777655543333333333221 PP
TCP 67 sssaseceaesssssasnsssg.........k 89
as+sss+
Araip.EDM7N 154 ---------------ASSSSSSrflkgkdlmT 170
...............22222222222211210 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 3.8E-26 | 57 | 170 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 28.442 | 57 | 115 | IPR017887 | Transcription factor TCP subgroup |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0030154 | Biological Process | cell differentiation | ||||
| GO:0045962 | Biological Process | positive regulation of development, heterochronic | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 256 aa Download sequence Send to blast |
MMMETKEEGM IIEENKKKSN ENDKFTLKGS AAASSSTRQW SAFRNPRIVR VSRSFGGKDR 60 HSKVCTIRGL RDRRIRLSVP TAIQLYDLQD KLGLSQPSKV IDWLLDATKL DIDKLPPLHF 120 PQPCFQPPQT LFPHLLHDPS SFWKPRFWEL DSSASSSSSS RFLKGKDLMT TMPSDNNNNK 180 AKWIFKSNNA DHENQEFGGG YTTQRLFPME TQTTTNNNPF QMGDYYGSYP MMNNGGLMQL 240 NHLSSLQVGP SGSSQL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zkt_A | 4e-18 | 62 | 116 | 1 | 55 | Putative transcription factor PCF6 |
| 5zkt_B | 4e-18 | 62 | 116 | 1 | 55 | Putative transcription factor PCF6 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Plays a pivotal role in the control of morphogenesis of shoot organs by negatively regulating the expression of boundary-specific genes such as CUC genes, probably through the induction of miRNA (e.g. miR164). Participates in ovule develpment (PubMed:25378179). {ECO:0000269|PubMed:17307931, ECO:0000269|PubMed:25378179}. | |||||
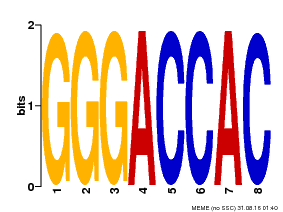
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00008 | PBM | Transfer from 496250 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.EDM7N |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020970331.1 | 1e-176 | transcription factor TCP5-like, partial | ||||
| Swissprot | Q9FME3 | 1e-49 | TCP5_ARATH; Transcription factor TCP5 | ||||
| TrEMBL | A0A444X7J2 | 1e-173 | A0A444X7J2_ARAHY; Uncharacterized protein | ||||
| STRING | GLYMA04G32324.1 | 2e-66 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF932 | 34 | 119 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G60970.1 | 6e-50 | TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



