 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.EMV9L | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 390aa MW: 43944.8 Da PI: 7.3277 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 54.6 | 2.4e-17 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g W++eEde+l++ ++++G g+W+++++ g+ R++k+c++rw +yl
Araip.EMV9L 14 KGLWSPEEDEKLLRHITKYGHGCWSSVPKQAGLQRCGKSCRLRWINYL 61
678*******************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 46.8 | 7.1e-15 | 67 | 110 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg +++eE+ l++++++ lG++ W+ Ia+ ++ gRt++++k+ w++
Araip.EMV9L 67 RGTFSQEEENLIIELHAVLGNR-WSQIAAQLP-GRTDNEIKNLWNS 110
899*******************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.8E-28 | 6 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 18.926 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.43E-29 | 12 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.7E-13 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.8E-15 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.95E-11 | 17 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 25.948 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.8E-26 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.8E-13 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 9.1E-14 | 67 | 111 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.11E-9 | 69 | 109 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001944 | Biological Process | vasculature development | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0010089 | Biological Process | xylem development | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010214 | Biological Process | seed coat development | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 390 aa Download sequence Send to blast |
MGRHSCCYKQ KLRKGLWSPE EDEKLLRHIT KYGHGCWSSV PKQAGLQRCG KSCRLRWINY 60 LRPDLKRGTF SQEEENLIIE LHAVLGNRWS QIAAQLPGRT DNEIKNLWNS CLKKKLRQRG 120 IDPVTHKPLS ELSEEKSQIQ EKALSNDNEL NLLRSESSNK SDGASSYDHQ QQQQQQQQQG 180 FVPTSEMEGS SSTNKDIFLD TTTRFMANTC SDFMGNYNMS YASSSSNVTV TDNNWFTQTS 240 RPSFDINSDF TSSSSFLPGS FCYKPESLKN TNMLSSSSSS WGLITDCSME EAEEAKWSEY 300 LHNQMLMLAA VQNNNNNNNN NNQASLCNEI KPLPSTTTVT TTPPPHLVPD TLGAIMLPHS 360 KNQHHHTSQT SSTTTFSKDI QNLTAAFGHI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 1e-28 | 12 | 116 | 5 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 111 | 117 | LKKKLRQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00134 | DAP | Transfer from AT1G09540 | Download |
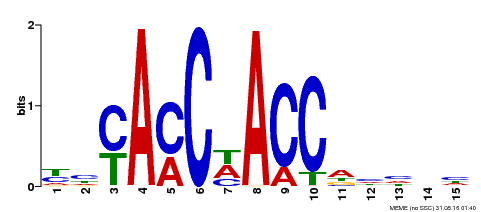
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.EMV9L |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT097326 | 1e-121 | BT097326.1 Soybean clone JCVI-FLGm-21J24 unknown mRNA. | |||
| GenBank | KT031185 | 1e-121 | KT031185.1 Glycine max clone HN_CCL_13 MYB/HD-like transcription factor (Glyma03g38410.1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016205675.1 | 0.0 | transcription repressor MYB6 | ||||
| Refseq | XP_025662903.1 | 0.0 | transcription repressor MYB6 | ||||
| TrEMBL | A0A444YJ20 | 0.0 | A0A444YJ20_ARAHY; Uncharacterized protein | ||||
| STRING | XP_007163255.1 | 1e-136 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1083 | 33 | 103 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G01680.3 | 5e-92 | myb domain protein 55 | ||||



