 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.HWR4Z | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 554aa MW: 61700.1 Da PI: 8.394 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 37.9 | 3.1e-12 | 388 | 434 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ +I++Lq
Araip.HWR4Z 388 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAIAHINELQ 434
799***********************66......******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 1.3E-25 | 35 | 161 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| Pfam | PF14215 | 1.0E-8 | 163 | 201 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.672 | 384 | 433 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 7.59E-18 | 387 | 450 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.98E-14 | 387 | 438 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 4.2E-17 | 388 | 450 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.0E-9 | 388 | 434 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 4.5E-16 | 390 | 439 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 554 aa Download sequence Send to blast |
MVMKILGKRA FDYLASNSFI TNESMLMATG SFENLQNKLS DLVERPNLNH FSWNYAIFWQ 60 FSQSKFKKDC VVLGWGDGCC REPIEGEEER EALRLGFDDD EVVQRMRKRV LQKLHTIFNG 120 SEEENENYAF GLDRVTDTEM FFLASMYFSF PKGYGGPGKC FELAGIKTVV LVPTELGVVE 180 LGSVRIVDEN LDLLKAVYSV FSSSFAHSSS FNDVGVPVPK IFGKELNVGN LNSGGKNFRE 240 KVVVRKIEEK KSWNGYPNGN NGIRFPNNAR NGVNGSSWAV NQGLSQGCLG GVFPPIPRSC 300 LSNMAKQADC GGKKDSLLEK FQRPPQGQVP MQIDFGVGSQ SFGRSIVGEC EISNSDSLHK 360 EEKPSATQEK RPKKRGRKPA NGREEPLNHV EAERQRREKL NQRFYALRAV VPNISKMDKA 420 SLLGDAIAHI NELQAKLKAL EYEKSTLKST SKANLSGIEA NLRDRTCSFK VDIEADQDGV 480 IVKVSCPIEL HPTSKLIQAL KDSEMSILES TFNATNDHVF HTFVIKSHAS EQLTKEKVIA 540 AISGESNSRK LIFA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 2e-29 | 382 | 449 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_B | 2e-29 | 382 | 449 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_E | 2e-29 | 382 | 449 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_F | 2e-29 | 382 | 449 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_G | 2e-29 | 382 | 449 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_I | 2e-29 | 382 | 449 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_M | 2e-29 | 382 | 449 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_N | 2e-29 | 382 | 449 | 2 | 69 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 369 | 377 | KRPKKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that negatively regulates jasmonate (JA) signaling (PubMed:30610166). Negatively regulates JA-dependent response to wounding, JA-induced expression of defense genes, JA-dependent responses against herbivorous insects, and JA-dependent resistance against Botrytis cinerea infection (PubMed:30610166). Plays a positive role in resistance against the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (PubMed:30610166). {ECO:0000269|PubMed:30610166}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00100 | PBM | Transfer from AT1G01260 | Download |
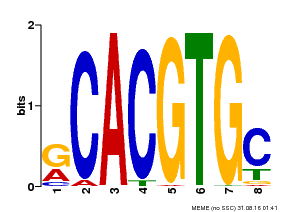
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.HWR4Z |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by wounding, feeding with herbivorous insects, infection with the fungal pathogen Botrytis cinerea and infection with the bacterial pathogen Pseudomonas syringae pv tomato DC3000. {ECO:0000269|PubMed:30610166}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016181554.1 | 0.0 | transcription factor bHLH13-like | ||||
| Swissprot | A0A3Q7ELQ2 | 1e-177 | MTB1_SOLLC; Transcription factor MTB1 | ||||
| TrEMBL | A0A445ACC5 | 0.0 | A0A445ACC5_ARAHY; Uncharacterized protein | ||||
| STRING | GLYMA19G44570.2 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5448 | 32 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 1e-124 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



