 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.JQQ8M | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 947aa MW: 105151 Da PI: 6.3225 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56.1 | 8.5e-18 | 38 | 84 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+e de+l +av+ + g++Wk+Ia+++ Rt+ qc +rwqk+l
Araip.JQQ8M 38 KGNWTPEQDEILRKAVQHFKGKNWKKIAECFK-DRTDVQCLHRWQKVL 84
79*****************************9.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 63.1 | 5.5e-20 | 90 | 136 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+W++eEde ++++v+++G++ W+tIa++++ gR +kqc++rw+++l
Araip.JQQ8M 90 KGPWSKEEDEVIIELVNKYGPKKWSTIAHHLP-GRIGKQCRERWHNHL 136
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 48.8 | 1.6e-15 | 142 | 184 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ +WT+eE++ l++a++ +G++ W+ + ++ gRt++++k++w+
Araip.JQQ8M 142 KEAWTQEEELTLIRAHQIYGNK-WAELTKFLP-GRTDNSIKNHWH 184
579*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.719 | 33 | 84 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 5.24E-15 | 36 | 91 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.9E-15 | 37 | 86 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.5E-15 | 38 | 84 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.7E-21 | 39 | 96 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.11E-13 | 40 | 84 | No hit | No description |
| PROSITE profile | PS51294 | 33.433 | 85 | 140 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.27E-31 | 87 | 183 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.7E-19 | 89 | 138 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.6E-18 | 90 | 136 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.93E-17 | 92 | 136 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-27 | 97 | 144 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 19.641 | 141 | 191 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.5E-14 | 141 | 189 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 9.1E-14 | 142 | 184 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.81E-11 | 144 | 184 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.5E-21 | 145 | 191 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032875 | Biological Process | regulation of DNA endoreduplication | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 947 aa Download sequence Send to blast |
MNGDRKIPVS PSKEQIDGAQ KIRALHGRTT GPTRRSTKGN WTPEQDEILR KAVQHFKGKN 60 WKKIAECFKD RTDVQCLHRW QKVLNPELVK GPWSKEEDEV IIELVNKYGP KKWSTIAHHL 120 PGRIGKQCRE RWHNHLNPGI NKEAWTQEEE LTLIRAHQIY GNKWAELTKF LPGRTDNSIK 180 NHWHSSVKKK LDSYLASGLL AQFQSAPLAG NPDQQMVLTS EMHSVGDGNG PKRTDREDVS 240 ECSQESADSA GLTSNVDLQT REVYKPEEES ILGKDHNLSQ ASCPQHYYIS LDDVDICIPE 300 VACQDACCSQ FAEQICSHKS GNALTGDYQF NCVDNSEMCN MVNVPFQTEG LGISTLMRPA 360 SMDPAKPNNM SISEDECCRF LFSEAVSDGC FSPRVYIKDM DMVDFSGCNM FLCQSCHIQI 420 PSPRESSSTS QLTCPKCSNN FKGSSSSQSF PPVFSANSEV KQEQPETPKD SSKLVPVNSF 480 GYGSNSKVTC YAKYGKLNEH SKKEDTGTLC YQPPCFPSLD IPFLSCDLAQ SEPDMQQEFS 540 PLGIRQLLMS PVTCLTPFRL WDSPSGGDNL DALFKSAAKT FTGTPSILSK RHRDLLSPLS 600 DKRTDKKLVT DMPSSLIKSF SSLDFMFDDN ENREADMISP TSLQKWSIAV SVDDDKENCG 660 QTVKDKQVEE KNNSAISDEK NRENGTVKNS SQDSLKHQVE QPSGILVERN MNDIAPNSPG 720 SDTVLGSSAR TPKNLSNRRL EAIPNQRSPC SRVKCVRAKE HEKLSVDVTC VGTTSGKQAK 780 NDGGFETSSI FGDTPFMKSI KSPSAWKSPW FINATFLCSP RIATEISIED YGYLMSSGNR 840 SYDAIGLMKQ ISEKTAATYA NAQEVLENET PEALPKNASL NDEEGGQDDN HLQKQSAKHS 900 QAASNALMER RILDFSECET TPSKGDNSKS SPVNFSSPSS YLLKGCR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 6e-67 | 38 | 191 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 6e-67 | 38 | 191 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00505 | DAP | Transfer from AT5G11510 | Download |
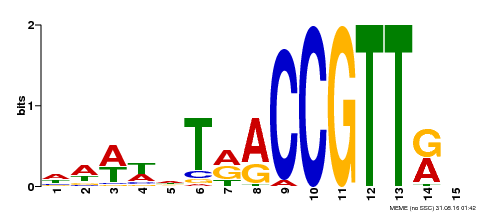
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.JQQ8M |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015035 | 8e-34 | AP015035.1 Vigna angularis var. angularis DNA, chromosome 2, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016207096.1 | 0.0 | transcription factor MYB3R-1 | ||||
| TrEMBL | A0A445CVN9 | 0.0 | A0A445CVN9_ARAHY; Uncharacterized protein | ||||
| STRING | GLYMA06G08660.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3727 | 33 | 62 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G11510.1 | 0.0 | myb domain protein 3r-4 | ||||



