 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.KHB2Z | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 327aa MW: 37417.5 Da PI: 5.9707 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 55.9 | 9.7e-18 | 29 | 76 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+WT +Ed+ l+++++ +G g+W++ ar g++Rt+k+c++rw++yl
Araip.KHB2Z 29 RGPWTVDEDLTLINYIATHGEGRWNSLARSAGLKRTGKSCRLRWLNYL 76
89********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 51.8 | 1.8e-16 | 82 | 125 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg+ T eE++l++++++++G++ W++Ia++++ gRt++++k++w++
Araip.KHB2Z 82 RGNITLEEQLLILELHTRWGNR-WSKIAQYLP-GRTDNEIKNYWRT 125
7999******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 23.849 | 24 | 80 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.57E-30 | 26 | 123 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.4E-15 | 28 | 78 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.1E-16 | 29 | 76 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.7E-22 | 30 | 83 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.23E-11 | 31 | 76 | No hit | No description |
| PROSITE profile | PS51294 | 18.099 | 81 | 131 | IPR017930 | Myb domain |
| SMART | SM00717 | 9.1E-14 | 81 | 129 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.0E-14 | 82 | 125 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.3E-22 | 84 | 130 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.23E-7 | 102 | 125 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009620 | Biological Process | response to fungus | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 327 aa Download sequence Send to blast |
MEVKGSSNSN NSAVLISHSE EEEMMDLRRG PWTVDEDLTL INYIATHGEG RWNSLARSAG 60 LKRTGKSCRL RWLNYLRPDV RRGNITLEEQ LLILELHTRW GNRWSKIAQY LPGRTDNEIK 120 NYWRTRVQKH AKQLKCDVNS KQFKDAMRYL WMPRLVERIQ AAAAASTTTT TVVAPTNTSI 180 NNNNNNAYTY NNNNLNNNFE VHNGKMMLTT TPPPSVMANN DFVGSQITTQ SYNTPENSST 240 AASSDSFGTP ISDFNDYYNS NNINNLGDNN FYQASQTLFS HEQGMDLQSM MMVESNTPWM 300 QSGDTSDNFW NVENMLFLEQ QLMNDNM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 1e-25 | 26 | 123 | 24 | 120 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00334 | DAP | Transfer from AT3G06490 | Download |
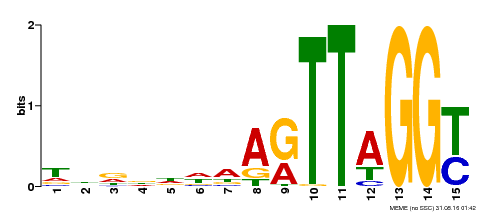
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.KHB2Z |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016177884.1 | 0.0 | transcription factor MYB108-like | ||||
| Refseq | XP_025630250.1 | 0.0 | transcription factor MYB108 | ||||
| TrEMBL | A0A445AW92 | 0.0 | A0A445AW92_ARAHY; Uncharacterized protein | ||||
| STRING | GLYMA17G04171.1 | 1e-129 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF875 | 34 | 125 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G49620.1 | 1e-86 | myb domain protein 78 | ||||



