 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.KK7TK | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | GRAS | ||||||||
| Protein Properties | Length: 519aa MW: 56189.2 Da PI: 4.7403 | ||||||||
| Description | GRAS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GRAS | 333.1 | 5.2e-102 | 207 | 469 | 1 | 277 |
GRAS 1 lvelLlecAeavssgdlelaqalLarlselaspdgdpmqRlaayfteALaarlarsvselykalppsetseknsseelaalklfsevsPilkfshlta 98
lv++Ll+cAeav++++l+la+al++++ la +++ +m+++a+yf++ALa+r+++ + p+e+ +++ s++l++ f+e +P+lkf+h+ta
Araip.KK7TK 207 LVHTLLACAEAVQQENLKLADALVKHVGLLAQSQAGAMKKVASYFAQALARRIYQ--------IYPQEAMDSSFSDVLHM--HFYESCPYLKFAHFTA 294
689***************************************************9........55677766667777776..5*************** PP
GRAS 99 NqaIleavegeervHiiDfdisqGlQWpaLlqaLasRpegppslRiTgvgspesgskeeleetgerLakfAeelgvpfefnvlvakrledleleeLrv 196
NqaIlea+++++rvH+iDf+++qG+QWpaLlqaL+ Rp+gpp++R+Tg+g+p+++++++l+++g++La++A+++gv+fef+ +v+++l+dl++e+L++
Araip.KK7TK 295 NQAILEAFASATRVHVIDFGLKQGMQWPALLQALTLRPGGPPTFRLTGIGPPQTDNTDSLQQVGWKLAQLAQTIGVQFEFRGFVCSSLSDLDTEMLEI 392
************************************************************************************************** PP
GRAS 197 kpgEalaVnlvlqlhrlldesvsleserdevLklvkslsPkvvvvveqeadhnsesFlerflealeyysalfdsleaklpr 277
+pgEa+aVn+v++lhr+l++s+s+e+ vL +vk+++Pk+++vveqea+hn++ F++rf+eal+yys+lfdsle ++++
Araip.KK7TK 393 RPGEAVAVNSVFELHRMLARSGSIEK----VLGTVKKIKPKIFTVVEQEANHNGPVFVDRFTEALHYYSSLFDSLEGSVAA 469
**************************....**********************************************99875 PP
| |||||||
| 2 | GRAS | 21 | 3.6e-07 | 486 | 509 | 351 | 374 |
GRAS 351 veeesgslvlgWkdrpLvsvSaWr 374
vee++g+l+lgW++r+L+++SaW+
Araip.KK7TK 486 VEENNGCLMLGWHTRSLIATSAWK 509
799********************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01129 | 3.4E-37 | 54 | 127 | No hit | No description |
| Pfam | PF12041 | 4.4E-36 | 54 | 121 | IPR021914 | Transcriptional factor DELLA, N-terminal |
| PROSITE profile | PS50985 | 55.769 | 181 | 519 | IPR005202 | Transcription factor GRAS |
| Pfam | PF03514 | 1.8E-99 | 207 | 469 | IPR005202 | Transcription factor GRAS |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042176 | Biological Process | regulation of protein catabolic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:2000033 | Biological Process | regulation of seed dormancy process | ||||
| GO:2000377 | Biological Process | regulation of reactive oxygen species metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 519 aa Download sequence Send to blast |
MKRDHSEAGS SYGQQGSSSS SVAKGECSSM SSGKAKMWEE DHDPSSGGGG GGMDELLAAL 60 GYKVRNSDMA DVAQKLEQLE MVMGTAQEDG ISHLATDTVH YDPTDLYSWV QSMINELNPP 120 PTTAAANGTS SSLLCSSQIE ENTSRAFTDD SEYDLRAIPG IAVYPQNDEP SNKRLKTNPV 180 PEIGSEGVQE PSRPVVLVDS QETGVRLVHT LLACAEAVQQ ENLKLADALV KHVGLLAQSQ 240 AGAMKKVASY FAQALARRIY QIYPQEAMDS SFSDVLHMHF YESCPYLKFA HFTANQAILE 300 AFASATRVHV IDFGLKQGMQ WPALLQALTL RPGGPPTFRL TGIGPPQTDN TDSLQQVGWK 360 LAQLAQTIGV QFEFRGFVCS SLSDLDTEML EIRPGEAVAV NSVFELHRML ARSGSIEKVL 420 GTVKKIKPKI FTVVEQEANH NGPVFVDRFT EALHYYSSLF DSLEGSVAAG LAAPPPTEDL 480 LMSELVEENN GCLMLGWHTR SLIATSAWKL PPLSSGGES |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3g_A | 7e-51 | 214 | 466 | 26 | 280 | Protein SCARECROW |
| 5b3h_A | 6e-51 | 214 | 466 | 25 | 279 | Protein SCARECROW |
| 5b3h_D | 6e-51 | 214 | 466 | 25 | 279 | Protein SCARECROW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional regulator that acts as a repressor of the gibberellin (GA) signaling pathway. Probably acts by participating in large multiprotein complexes that represses transcription of GA-inducible genes. Upon GA application, it is degraded by the proteasome, allowing the GA signaling pathway (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00611 | ChIP-seq | Transfer from AT2G01570 | Download |
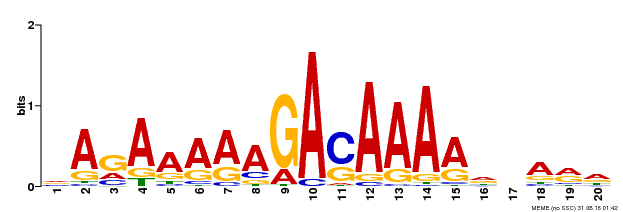
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.KK7TK |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020975220.1 | 0.0 | LOW QUALITY PROTEIN: DELLA protein GAI | ||||
| Swissprot | Q84TQ7 | 0.0 | GAI_GOSHI; DELLA protein GAI | ||||
| TrEMBL | A0A444ZVZ0 | 0.0 | A0A444ZVZ0_ARAHY; Uncharacterized protein | ||||
| STRING | XP_007163387.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF803 | 34 | 121 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G03450.1 | 0.0 | RGA-like 2 | ||||



