 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.LQ8SS | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1032aa MW: 114212 Da PI: 8.397 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 128.8 | 2e-40 | 118 | 195 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C++dls+ak+yhrrhkvCe hska+++ + +++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+q+
Araip.LQ8SS 118 MCQVDNCKEDLSSAKDYHRRHKVCELHSKASKAPLGNQMQRFCQQCSRFHPLTEFDEGKRSCRRRLAGHNRRRRKTQP 195
6**************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 7.9E-34 | 113 | 180 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.641 | 116 | 193 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 3.14E-37 | 117 | 197 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 5.7E-30 | 119 | 192 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 5.3E-5 | 819 | 915 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 9.02E-6 | 820 | 916 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.32E-4 | 824 | 916 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1032 aa Download sequence Send to blast |
MEKVAPPIFM HQALQSRFCD VPMTTTATTK KRDLSYDVDK SSSNGNWNPN AWSWDSVRFI 60 GKPVPQQNGV VSVEEETLRL NLGSSGVSGS GGSVDPSVSR PSKRVRSGSP TGTASYPMCQ 120 VDNCKEDLSS AKDYHRRHKV CELHSKASKA PLGNQMQRFC QQCSRFHPLT EFDEGKRSCR 180 RRLAGHNRRR RKTQPEDVTS QPESVATGNA EIFNLLSAIA SSQGQKPNGK FEDRSKIASQ 240 VPDKDQLVQI LNRIPLPADL AAKLLNVGGK GQIQTSSYHH DKVNQSNSGP LTKDLLAVLS 300 TTLSASTPNS QKSSQSSDSE KSRASADQVG ESLQMRQYPQ EFASVGDERS SGSSQSPVED 360 SDFPEVRVNL PLQLFSSSPE VGNPPKLTPS QKYFSSDSSN PVEERSPSSS PAVDNQFDLQ 420 GVARGLKADG VPSRREVNAN KEASQSQSYN ISLNLFNAPN SRVQPSSLQS VPFQAGYASS 480 GSDHSPPSLN SDAQDRTGRI MFKLFDKDPS HFPGTLRTQI YNWLSNSPSD MESYIRPGCV 540 VLSLYASMSS AAWEQLEENF LQHVHSLIQS NSDFWRIGRF LVHSGNQLAL HKDGKIHLCK 600 PWRTWRSPEL ISVSPLAIVS GQETSFSLKG RNLSNPGTKI HCTGTGGYTP IKVVESACYG 660 MTYDKIKLSG IKVQDASPGL LGRCFIEVEN GFKGSSFPVI IADATICKEL RPLESEFDKE 720 ENASDAISDE HGYDLGRPRS REETLHFLNE LGWLFQRKRF SYTDLVPDYS LDRFRFILTF 780 SVERNCCMLV KTLLDMLVQK YLEGQWSSTA SLEMLNAIQL LNRAVKRKYV NMVDLLIQYA 840 VPSNNDASRK YVFPPNVAGP DGITPLHLAA CTSSSEGVID SLTNDPQEIG LNSWDSLLDA 900 NGQTPHAYAM MRNNHSYNVL VARKLSNRRR GHVSVTINSE IEHSSMDIEL KQRQSDQTKR 960 GQNSCTKCSA MADVRYSSRI PGSRSFGHHR PFIHSILAIA AVCVCVCLFL RGHPWVGSVT 1020 PFNWEKLDYG TI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 3e-31 | 110 | 192 | 2 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
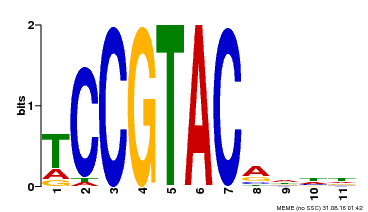
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.LQ8SS |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF312233 | 0.0 | KF312233.1 Arachis hypogaea squamosa promoter-binding-like protein (SPL14) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016180086.1 | 0.0 | squamosa promoter-binding-like protein 14 isoform X1 | ||||
| Refseq | XP_025682262.1 | 0.0 | squamosa promoter-binding-like protein 14 isoform X1 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | A0A444XAR9 | 0.0 | A0A444XAR9_ARAHY; Uncharacterized protein | ||||
| STRING | GLYMA20G00900.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF9694 | 31 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



