 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.P9FMN | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 585aa MW: 66138.4 Da PI: 7.98 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 86.7 | 3.4e-27 | 111 | 213 | 1 | 91 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkk............tekerrtraetrtgCkaklkvkkekdgkwevtkleleHn 86
+fY+eYA+++GF++ +++s++sk+++e+++++f Cs++g+++e +k+ +e++ +r+ ++t+Cka+++vk+++dgkw+++++++eHn
Araip.P9FMN 111 SFYQEYARSMGFNTAIQNSRRSKTSREFIDAKFACSRYGTKREYDKSfnrprarqskpdSENSTGRRSCSKTDCKASMHVKRKHDGKWVIHSFVKEHN 208
5********************************************999***99999987777777999****************************** PP
FAR1 87 Helap 91
Hel p
Araip.P9FMN 209 HELLP 213
**975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 4.5E-25 | 111 | 213 | IPR004330 | FAR1 DNA binding domain |
| PROSITE profile | PS50966 | 9.681 | 330 | 366 | IPR007527 | Zinc finger, SWIM-type |
| Pfam | PF04434 | 1.3E-5 | 337 | 364 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 2.3E-6 | 341 | 368 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 585 aa Download sequence Send to blast |
MASVTAAICL TLHIIKKKAV LNMDIDLRLP SGEHDKEDEE TATIDNILDG EEKLNNGAME 60 GRNMVDPGME LQALNGGDLN SPTLDMVIFK EDTNLEPLSG MEFESHGEAY SFYQEYARSM 120 GFNTAIQNSR RSKTSREFID AKFACSRYGT KREYDKSFNR PRARQSKPDS ENSTGRRSCS 180 KTDCKASMHV KRKHDGKWVI HSFVKEHNHE LLPAQAVSEQ TRRMYAAMAR QFAEYKTVTS 240 VQDFVKLYEA ILQDRYEEEA KADSDTWNKV ATLKTPSPLE KSVAGVYTHA VFKKIQAEVV 300 GAVACHPKAD RQDETTIIHR VHDMQSNKDF FVVVNQVKSE WSCICRLFEY RGYLCRHILI 360 VLQYSGHSAI PSHYILKRWT KDAKVRNIMG EESEHMLARV QRYNDLCQRA FKLSEEGSLS 420 QESYSIAVHA LHEAQKSCVS VNNSGKSPME VGTSVTHAQL STEEDTQSRN MGKSNKKKNP 480 AKKKKDKFST RAAVTLDGYY GTQQQVQGML NLMGPRDDFY GNQQTLQGLG PISSIPTSHD 540 GYYSAHQGMP NLAQLDFLRT GFTYGIRDDP NVRAAQLHED PSRHA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
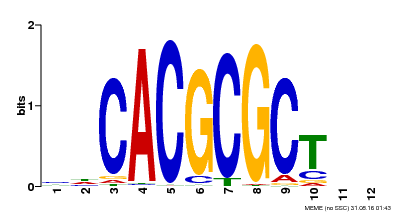
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. When associated with PHYA, protects it from being recognized and degraded by the COP1/SPA complex. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:17012604, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:18715961, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00078 | ChIP-seq | Transfer from AT3G22170 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.P9FMN |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. Up-regulated by white light. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015040 | 0.0 | AP015040.1 Vigna angularis var. angularis DNA, chromosome 7, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025624748.1 | 0.0 | protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X4 | ||||
| Swissprot | Q9LIE5 | 1e-122 | FHY3_ARATH; Protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| TrEMBL | A0A0A0KV81 | 0.0 | A0A0A0KV81_CUCSA; Uncharacterized protein | ||||
| STRING | XP_007148758.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF40 | 30 | 530 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22170.2 | 1e-120 | far-red elongated hypocotyls 3 | ||||



