 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.PU48P | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 838aa MW: 92220 Da PI: 5.7769 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 71.1 | 1.4e-22 | 656 | 752 | 2 | 95 |
EEE-..-HHHHTT-EE--HHH.HTT..---..--SEEEEEE.TTS-EEEEEE....EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SS.SEE..E CS
B3 2 fkvltpsdvlksgrlvlpkkfaeeh..ggkkeesktltled.esgrsWevkliy..rkksgryvltkGWkeFvkangLkegDfvvFkldgr.sefelv 93
+kvl++sdv+++gr+vlpkk ae+h +++ + ++t+t+ed ++r+W+++++y ++ks++y+l+ ++ +FvkangL+egDf+v++ ++ ++++++
Araip.PU48P 656 QKVLKQSDVGSLGRIVLPKKEAETHlpELEARDGITITMEDiGTSRVWNMRYRYwpNNKSRMYLLE-NTGDFVKANGLQEGDFIVIY--SDvKCGKFM 750
79*************************99999*********7778*********777777777777.********************..555899777 PP
EE CS
B3 94 vk 95
++
Araip.PU48P 751 IR 752
65 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 4.9E-30 | 648 | 763 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10015 | 8.58E-37 | 653 | 754 | No hit | No description |
| SuperFamily | SSF101936 | 2.75E-24 | 654 | 751 | IPR015300 | DNA-binding pseudobarrel domain |
| SMART | SM01019 | 3.2E-23 | 655 | 755 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 11.42 | 655 | 757 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 9.7E-19 | 656 | 752 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009657 | Biological Process | plastid organization | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0031930 | Biological Process | mitochondria-nucleus signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0001076 | Molecular Function | transcription factor activity, RNA polymerase II transcription factor binding | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 838 aa Download sequence Send to blast |
PSQRSSFVGV QGLDLHARVA KTETVDPILG FGAMDHHHHN NHNHHDAEIY ADADDADKDL 60 WLNTTTEQED LLVGDVKTDA SMFYDEFPPL PDFPCMSSSS SSSSTPTLPL KNLACSTTST 120 TTATAATSSS SSSASSWAVL KPEAEDNNNG EWNCSNKQQL YMQHHDPLDA TGALSSTASM 180 EISQPKFPDQ GGFDGVVGDV DCMDDVMDTF GYMELLEANE FFDPSSIFQG EENPLAEFTQ 240 EDQVLPQQEE QQQQEKPAVV HNQNDLIVLR PQQEAVVEGV SNNNYQQLLG LWEASNNNDA 300 EIQGDGGGGG AIADDEMSMV FLEWLKSNKD SVSASDLRSV KLKKSTIESA AMRLGGGKEA 360 MKQLLKLILE WVQTSHLQSR RRKENNNDNT VANPFTEQFQ NPIHQNNPNA SSSFAIEPNT 420 CFTQHQPWMD QVPLMAAAPP SQSQPQPQFQ QPTIGYVGDP YTNGASANTI NGSNNFQAGN 480 EYHMLESANS WPPSQYTVTP YYTQPFGDHS FQPPAAGGFG NQYPYQFFHG PGDGLIRLGP 540 SATKEARKKR MARQRRLSSH SRHHINLQSQ GSDPNTRLGV GGENCTGVVS AAPANWVYWP 600 TMAGAVASVA PVAPAEPSAV VDKPAMQTQN YHHQGRLSSD RRQGWKSEKN LRFLLQKVLK 660 QSDVGSLGRI VLPKKEAETH LPELEARDGI TITMEDIGTS RVWNMRYRYW PNNKSRMYLL 720 ENTGDFVKAN GLQEGDFIVI YSDVKCGKFM IRGVKVRQQE GAKAEGKKTG KSHKNQHGNN 780 AVPTAGEPHT NATSSSTHQE NEKSINNNII LCSRIKGTTL NTMHIISSVF LHGRTKLG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j9b_A | 4e-39 | 651 | 760 | 1 | 110 | B3 domain-containing transcription factor FUS3 |
| 6j9b_D | 4e-39 | 651 | 760 | 1 | 110 | B3 domain-containing transcription factor FUS3 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00083 | PBM | Transfer from AT3G24650 | Download |
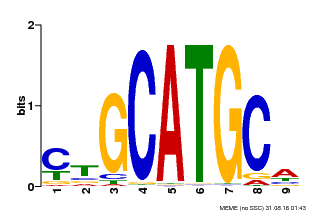
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.PU48P |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016198322.1 | 0.0 | B3 domain-containing transcription factor ABI3 isoform X2 | ||||
| TrEMBL | A0A444ZN17 | 0.0 | A0A444ZN17_ARAHY; Uncharacterized protein | ||||
| STRING | GLYMA08G47240.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF9448 | 30 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G24650.1 | 1e-126 | B3 family protein | ||||



